GenAge entry for BLM (Homo sapiens)
Gene name (HAGRID: 68)
- HGNC symbol
- BLM
- Aliases
- BS; RECQL3; RECQ2
- Common name
- Bloom syndrome, RecQ helicase-like
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
BLM is a helicase involved in DNA repair and replication. Mutations in BLM give rise to Bloom’s syndrome. Bloom’s syndrome cells are characterised by chromosomal aberrations including chromatid gaps and breaks, telomere association and quadriradial chromosomes, resulting from defects in DNA repair [4492]. Although Bloom’s syndrome is not considered accelerated ageing, BLM and WRN belong to the same family of proteins. Therefore, it is possible, even if unproven, that BLM plays a role in human ageing [444].
Cytogenetic information
- Cytogenetic band
- 15q26.1
- Location
- 90,717,326 bp to 90,815,462 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000079; regulation of cyclin-dependent protein serine/threonine kinase activity
GO:0000724; double-strand break repair via homologous recombination
GO:0000729; DNA double-strand break processing
GO:0000731; DNA synthesis involved in DNA repair
GO:0000732; strand displacement
GO:0000733; DNA strand renaturation
GO:0006260; DNA replication
GO:0006281; DNA repair
GO:0006310; DNA recombination
GO:0006974; cellular response to DNA damage stimulus
GO:0007095; mitotic G2 DNA damage checkpoint
GO:0010165; response to X-ray
GO:0016925; protein sumoylation
GO:0031297; replication fork processing
GO:0032508; DNA duplex unwinding
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045910; negative regulation of DNA recombination
GO:0048478; replication fork protection
GO:0051259; protein oligomerization
GO:0051782; negative regulation of cell division
GO:0071479; cellular response to ionizing radiation
GO:0072711; cellular response to hydroxyurea
GO:0072757; cellular response to camptothecin
GO:1901796; regulation of signal transduction by p53 class mediator
Cellular component: GO:0000228; nuclear chromosome
GO:0000781; chromosome, telomeric region
GO:0000800; lateral element
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005730; nucleolus
GO:0005737; cytoplasm
GO:0016363; nuclear matrix
GO:0016605; PML body
Hide GO termsFunction: GO:0000405; bubble DNA binding
GO:0002039; p53 binding
GO:0003697; single-stranded DNA binding
GO:0004003; ATP-dependent DNA helicase activity
GO:0004386; helicase activity
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0008026; ATP-dependent helicase activity
GO:0009378; four-way junction helicase activity
GO:0016887; ATPase activity
GO:0036310; annealing helicase activity
GO:0043140; ATP-dependent 3'-5' DNA helicase activity
GO:0051880; G-quadruplex DNA binding
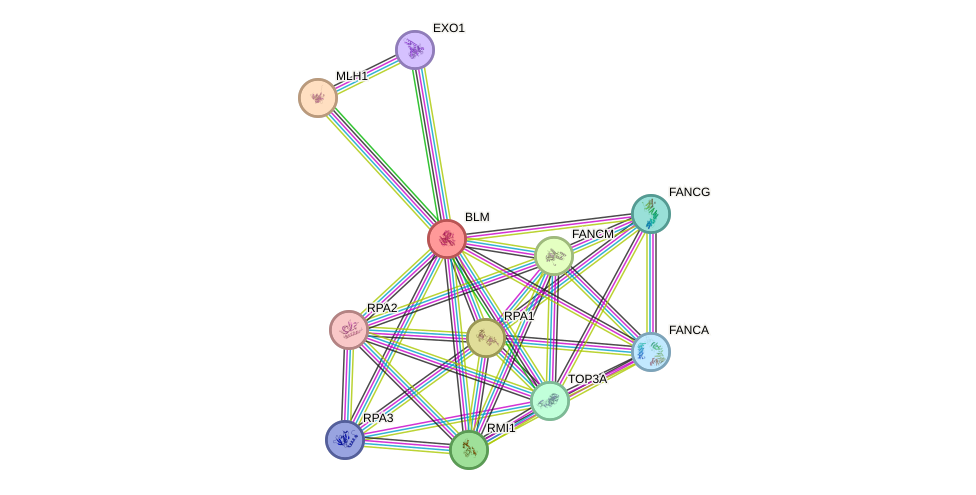
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, ATM, WRN, EGFR, NBN, PARP1, BRCA1, RPA1, BLM, VCP, HSP90AA1, TOP2A, RAD51, UBE2I, TERF1, FEN1, TERF2, SUMO1, H2AFX, ATR, MLH1, SIRT7, TP53BP1
- STRING interaction network
Retrieve sequences for BLM
Homologs in model organisms
- Danio rerio
- blm
- Drosophila melanogaster
- mus309
- Mus musculus
- Blm
- Rattus norvegicus
- Blm
- Schizosaccharomyces pombe
- rqh1