GenAge entry for POLB (Homo sapiens)
Gene name (HAGRID: 236)
- HGNC symbol
- POLB
- Aliases
- Common name
- polymerase (DNA directed), beta
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
POLB is involved in DNA maintenance, replication, and recombination. It appears to play a role in DNA repair and it interacts with WRN [1520]. In mice, POLB haploinsufficiency increases cancer risk with age. While lifespan is not significantly altered, haploinsufficient mice may exhibit an increase in age-related mortality [1815]. POLB's role, if any, in human ageing remains to be determined.
Cytogenetic information
- Cytogenetic band
- 8p11.2
- Location
- 42,338,455 bp to 42,371,813 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0006261; DNA-dependent DNA replication
GO:0006281; DNA repair
GO:0006284; base-excision repair
GO:0006286; base-excision repair, base-free sugar-phosphate removal
GO:0006287; base-excision repair, gap-filling
GO:0006288; base-excision repair, DNA ligation
GO:0006290; pyrimidine dimer repair
GO:0006297; nucleotide-excision repair, DNA gap filling
GO:0006954; inflammatory response
GO:0006974; cellular response to DNA damage stimulus
GO:0007435; salivary gland morphogenesis
GO:0007568; aging
GO:0008630; intrinsic apoptotic signaling pathway in response to DNA damage
GO:0010332; response to gamma radiation
GO:0016446; somatic hypermutation of immunoglobulin genes
GO:0045471; response to ethanol
GO:0048535; lymph node development
GO:0048536; spleen development
GO:0048872; homeostasis of number of cells
GO:0051402; neuron apoptotic process
GO:0055093; response to hyperoxia
GO:0071707; immunoglobulin heavy chain V-D-J recombination
GO:0071897; DNA biosynthetic process
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005874; microtubule
GO:0005876; spindle microtubule
GO:0043234; protein complex
Hide GO termsFunction: GO:0003684; damaged DNA binding
GO:0003887; DNA-directed DNA polymerase activity
GO:0003906; DNA-(apurinic or apyrimidinic site) lyase activity
GO:0005515; protein binding
GO:0008017; microtubule binding
GO:0016829; lyase activity
GO:0019899; enzyme binding
GO:0046872; metal ion binding
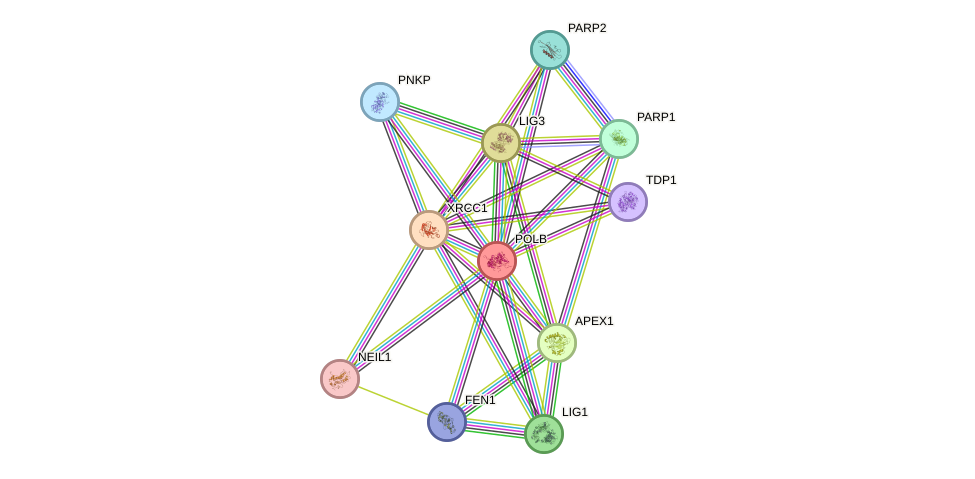
Protein interactions and network
- Protein-protein interacting partners in GenAge
- WRN, PARP1, EP300, APTX, PCNA, APEX1, STUB1, TPP2
- STRING interaction network
Retrieve sequences for POLB
Homologs in model organisms
In other databases
- LongevityMap
- This gene is present as POLB