The Evolution of Ageing
Species differences in longevity (e.g., why does a mouse cannot live more than 5 years yet humans can live over 100?) remain a major puzzle of biology. The data in our AnAge database is ideal to study the evolution of longevity and ageing in vertebrate lineages. Our aim in this project is therefore to study the events, including genetic changes, shaping longevity in different lineages to help understand why different species age at different paces. As an example of longevity differences between species, below is the phylogenetic tree of mammals with the average maximum longevity (in blue) of each major mammalian clade.
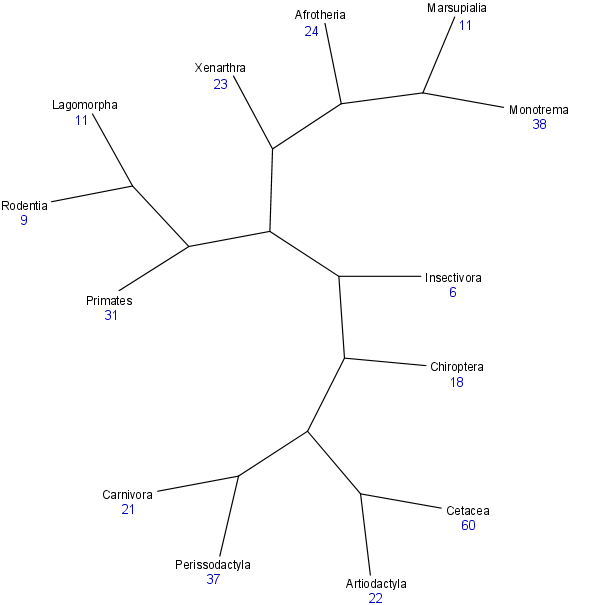
Legend: Phylogeny based on published results. Figure rendered using TreeView. Average longevity for major mammalian taxa calculated from AnAge built 9. Branch lengths are not to scale.
From the figure above, it is clear that longevity evolved differently in different mammalian clades. In other words, there is a phylogenetic effect on longevity, though potential sources of bias, such as body weight, must also be considered. Our goal then is to use this type of analysis to identify phylogenetic effects on longevity and also couple this information with genome and experimental data to identify molecular and genetic changes that may be responsible for the evolution of longevity.
Specifically, we are studying the molecular evolution of the ageing-related genes in GenAge across species with different rates of ageing. We initially performed such analysis using human-chimpanzee gene pairs and found that, surprisingly, ageing-related genes tend to evolve slower than average (other groups have since confirmed this observation). Another approach we have used so far involves the study of positions in the human genome that have been previously associated with human diseases. If the disease-causing positions are conserved in non-human primates this may help us understand the evolution of disease, longevity, and maybe even ageing. Because a limited number of mammalian genomes have been sequenced, by studying the mitochondrial genome we can obtain data from many more species. Therefore, we analysed the evolution of human disease-associated positions in the mitochondrial genome in primates.
As more mammalian genomes are sequenced, including by us, more detailed models of genome evolution can be derived to, for example, identify candidate longevity genes by searching for genes with unique signatures of selection in long-lived species. In this context, we developed a method to identify candidate genes involved in species differences in ageing based on detecting proteins with accelerated evolution in multiple lineages where longevity increased.
Lastly, we are also interesting in understanding differences between human populations and individuals. We developed a novel method to detect genes and processes in which populations most diverge which allowed us to detect novel candidates for contributing to population-specific traits.
As more genomes are sequenced, we want to further develop our methods and expand our analyses to gather more information about the evolution of longevity and ageing in vertebrates.