Integrative Systems Biology, Comparative Genomics, and Ageing
We developed the Human Ageing Genomic Resources (HAGR) to help understand how the genome regulates human ageing. Because ageing is a complex process involving the interplay of multiple genes and proteins with each other and with the environment, we believe that studying its multiple components as a whole is imperative to fully comprehend ageing and more accurately pinpoint how to intervene in it. HAGR was developed to provide the most accurate and complete information possible to permit such integrative analyses. While we cannot offer a full description of ageing, systems biology, or comparative genomics, we provide herein a brief description of our scientific strategy in order for users to better use the resources available in HAGR. For more information about ageing, we refer to our parent senescence.info website.
Arguably there are two key questions concerning ageing (de Magalhaes & Toussaint 2004): 1) What are the genetic determinants of ageing, both in terms of longevity differences between individuals and species differences in ageing? 2) Which changes occur across the lifetime to increase vulnerability, for example in a person from age 30 to age 70 to increase the chance of dying by roughly 30-fold (Figure 1)? HAGR was developed to facilitate studies that help answer both these questions. This is particularly timely because a variety of high-throughput technologies, including next-generation sequencing platforms (de Magalhaes et al. 2010), are now available that generate large amounts of data, which means there is a need to collect and systematically organize what we know about the genetics and genomics of ageing.
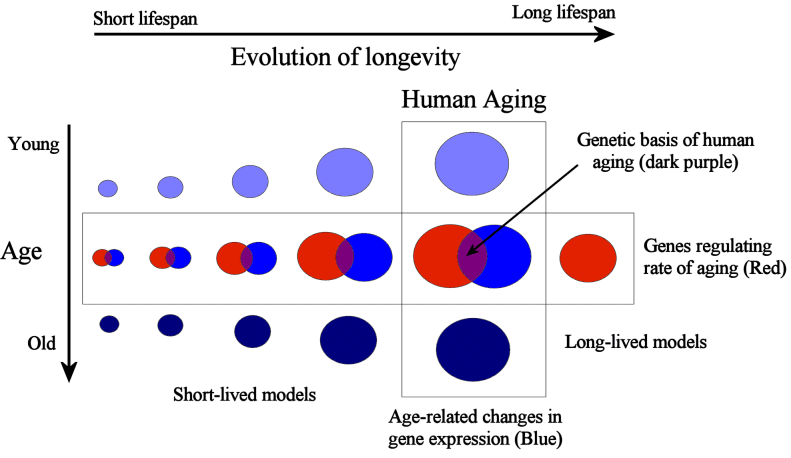
Figure 1: On one hand, comparative genomics can be used to study species differences in ageing. In parallel, we may study the changes people, or animals, endure while they age. These complementary approaches can help decipher the human ageing process and ultimately lead to interventions that extend life and health by manipulating ageing. Notice how the area of the circles decreases as we study species progressively more distant to humans, since it is expected that species evolutionary more distant from humans are less likely to share mechanisms of ageing that are relevant in humans.
One basic principle behind our approach is that the genome regulates rate of ageing in mammals, including humans, to a large extent (Miller 1999; de Magalhaes 2003). Therefore, in theory, it is possible to study how the human genome regulates ageing and age-related deterioration through computational approaches. The human genome is indecipherable by itself, however. To harness its information one powerful approach is comparative genomics (Ureta-Vidal et al. 2003). That is, we must compare the human genome to that of other organisms to understand which regions of the genome do what. As such, having fully sequenced genomes allows researchers to study the evolution of ageing with unprecedented detail. Even though the nature of the human ageing process remains unclear (de Magalhaes 2005), it is undeniable that certain genes make humans age slower than other primates and about 30 times slower than mice and rats. Finding those genes has tremendous biomedical applications and is one of the reasons why we created a database of ageing in animals to study the evolution of ageing. Following the same rationale, studying long-lived animals may allow us to identify adaptations that contribute to longevity and disease resistance (de Magalhaes 2006), and we are involved in genome sequencing and analysis projects.
Differences in longevity between individuals of the same species, including humans, are also important determinants of longevity and for this we developed the LongevityMap featuring genetic variants associated with human longevity. Moreover, we created GenAge, a database of genes related to ageing. Most researchers agree that ageing is a complex, multigenic process. GenAge allows us to focus on the genes and pathways more likely to be involved in ageing, in humans and in model organisms, and it allows us to study the interactions between the genes and how they together modulate longevity. Network analyses, in fact, are now an emerging paradigm to study how genes interact with each and with the environment to determine a whole phenotype.
In addition to understanding the genetic basis for phenotypic variation in ageing and longevity, it is also crucial to elucidate the changes that contribute to age-related degeneration. We have derived a common molecular signatures of ageing from gene expression data and made this available as part of GenAge. Moreover, our gene-centric databases, GenAge, GenDR and the LongevityMap, help interpret results from large-scale approaches, including gene expression profiling, to gain insights on the molecular drivers of the process of ageing.
In HAGR, we try interpret what we know about ageing in model organisms in light of human biology. Using a system-level approach that incorporates data from multiple sources, we attempt to build a more coherent model of the genetic and molecular mechanisms of human ageing (de Magalhaes & Toussaint 2004). Defining a gene as related to human ageing is subjective. We used different criteria to define different pathways and advise researchers to look at our gene database, as described elsewhere. By building and constantly upgrading GenAge we aim to provide resources and directions for research in biogerontology.
Bibliography and Recommended Reading
Austad, S. N. (2005) "Diverse aging rates in metazoans: targets for functional genomics." Mech Ageing Dev 126(1):43-49. PubMed
Charnov, E. L. (1993). Life History Invariants: Some Explorations of Symmetry in Evolutionary Ecology. Oxford University Press, Oxford.
D'Haeseleer, P., Liang, S., and Somogyi, R. (2000). "Genetic network inference: from co-expression clustering to reverse engineering." Bioinformatics 16(8):707-726. PubMed
de Magalhaes, J. P. (2003) "Is mammalian aging genetically controlled?" Biogerontology 4(2):119-120. PubMed
de Magalhaes, J. P., and Toussaint, O. (2004) "How bioinformatics can help reverse engineer human aging." Ageing Res Rev 3(2):125-141. PubMed
de Magalhaes, J. P. (2005) "Open-minded scepticism: inferring the causal mechanisms of human ageing from genetic perturbations." Ageing Res Rev 4(1):1-22. PubMed
de Magalhaes, J. P. (2006). "Species selection in comparative studies of aging and antiaging research." In: Handbook of Models for Human Aging, Conn, P. M. (ed.). Elsevier Academic Press, Burlington, MA, 9-20.
de Magalhaes, J. P. (2009). "Aging Research in the Post-Genome Era: New Technologies for an Old Problem." In: Redox Metabolism and Longevity Relationships in Animals and Plants, Foyer, C. H., Faragher, R. and Thornalley, P. J. (ed.). Taylor and Francis, New York and Abingdon, 99-115.
de Magalhaes, J. P., Finch, C. E., and Janssens, G. (2010). "Next-generation sequencing in aging research: emerging applications, problems, pitfalls and possible solutions." Ageing Res Rev 9(3):315-323. PubMed
Finch, C. E. (1990). Longevity, Senescence, and the Genome. The University of Chicago Press, Chicago and London.
Giallourakis, C., Henson, C., Reich, M., Xie, X., and Mootha, V. K. (2005) "Disease gene discovery through integrative genomics." Annu Rev Genomics Hum Genet 6:381-406. PubMed
Hedges, S. B. (2002) "The origin and evolution of model organisms." Nat Rev Genet 3(11):838-849. PubMed
Hood, L., and Galas, D. (2003). "The digital code of DNA." Nature 421(6921):444-448. PubMed
Ideker, T., Galitski, T., and Hood, L. (2001). "A new approach to decoding life: systems biology." Annu Rev Genomics Hum Genet 2:343-372. PubMed
Kitano, H. (2002). "Looking beyond the details: a rise in system-oriented approaches in genetics and molecular biology." Curr Genet 41(1):1-10. PubMed
Miller, R. A. (1999). "Kleemeier award lecture: are there genes for aging?" J Gerontol A Biol Sci Med Sci 54(7):B297-307. PubMed
Rose, M. R. (1991). Evolutionary Biology of Aging. Oxford University Press, New York.
Selinger, D. W., Wright, M. A., and Church, G. M. (2003). "On the complete determination of biological systems." Trends Biotechnol 21(6):251-254. PubMed
Ureta-Vidal, A., Ettwiller, L., and Birney, E. (2003). "Comparative genomics: genome-wide analysis in metazoan eukaryotes." Nat Rev Genet 4(4):251-262. PubMed