GenAge entry for BAK1 (Homo sapiens)
Gene name (HAGRID: 278)
- HGNC symbol
- BAK1
- Aliases
- BCL2L7; BAK; CDN1
- Common name
- BCL2-antagonist/killer 1
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
The BCL2 oncogene is a potent suppressor of apoptosis under diverse conditions. BAK1, a gene belonging to the BCL2 family, promotes cell death and counteracts the protection from apoptosis provided by BCL2 [2198]. Bak knockout mice showed reduced age-related apoptotic cell death of spiral ganglion neurons and hair cells in the cochlea, which in turn resulted in the prevention of age-related hearing loss [2200].
Cytogenetic information
- Cytogenetic band
- 6p21.3
- Location
- 33,572,546 bp to 33,580,293 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0001782; B cell homeostasis
GO:0001783; B cell apoptotic process
GO:0001836; release of cytochrome c from mitochondria
GO:0001974; blood vessel remodeling
GO:0002262; myeloid cell homeostasis
GO:0002352; B cell negative selection
GO:0006987; activation of signaling protein activity involved in unfolded protein response
GO:0007420; brain development
GO:0007568; aging
GO:0008053; mitochondrial fusion
GO:0008283; cell proliferation
GO:0008285; negative regulation of cell proliferation
GO:0008630; intrinsic apoptotic signaling pathway in response to DNA damage
GO:0008635; activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c
GO:0009620; response to fungus
GO:0010046; response to mycotoxin
GO:0010225; response to UV-C
GO:0010248; establishment or maintenance of transmembrane electrochemical gradient
GO:0010332; response to gamma radiation
GO:0010524; positive regulation of calcium ion transport into cytosol
GO:0010629; negative regulation of gene expression
GO:0014070; response to organic cyclic compound
GO:0019835; cytolysis
GO:0031018; endocrine pancreas development
GO:0031100; animal organ regeneration
GO:0032469; endoplasmic reticulum calcium ion homeostasis
GO:0032471; negative regulation of endoplasmic reticulum calcium ion concentration
GO:0033137; negative regulation of peptidyl-serine phosphorylation
GO:0034644; cellular response to UV
GO:0035108; limb morphogenesis
GO:0042493; response to drug
GO:0042542; response to hydrogen peroxide
GO:0043065; positive regulation of apoptotic process
GO:0043496; regulation of protein homodimerization activity
GO:0043497; regulation of protein heterodimerization activity
GO:0044346; fibroblast apoptotic process
GO:0045471; response to ethanol
GO:0045862; positive regulation of proteolysis
GO:0046902; regulation of mitochondrial membrane permeability
GO:0048597; post-embryonic camera-type eye morphogenesis
GO:0051726; regulation of cell cycle
GO:0051881; regulation of mitochondrial membrane potential
GO:0060068; vagina development
GO:0070059; intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress
GO:0070242; thymocyte apoptotic process
GO:0071260; cellular response to mechanical stimulus
GO:0090200; positive regulation of release of cytochrome c from mitochondria
GO:0097190; apoptotic signaling pathway
GO:0097192; extrinsic apoptotic signaling pathway in absence of ligand
GO:0097202; activation of cysteine-type endopeptidase activity
GO:1900103; positive regulation of endoplasmic reticulum unfolded protein response
GO:1901030; positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway
GO:1902262; apoptotic process involved in blood vessel morphogenesis
GO:1903896; positive regulation of IRE1-mediated unfolded protein response
Cellular component: GO:0005739; mitochondrion
GO:0005741; mitochondrial outer membrane
GO:0005783; endoplasmic reticulum
GO:0005829; cytosol
GO:0031307; integral component of mitochondrial outer membrane
GO:0046930; pore complex
Hide GO termsFunction: GO:0005515; protein binding
GO:0031072; heat shock protein binding
GO:0042802; identical protein binding
GO:0042803; protein homodimerization activity
GO:0044325; ion channel binding
GO:0046872; metal ion binding
GO:0046982; protein heterodimerization activity
GO:0051087; chaperone binding
GO:0051400; BH domain binding
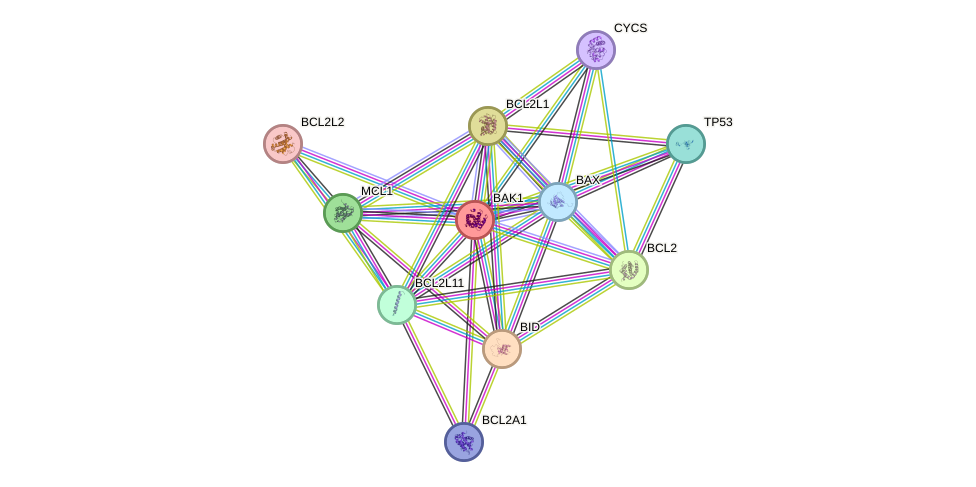
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, BCL2, BAX, HSPD1, DBN1, BAK1
- STRING interaction network