GenAge entry for HDAC3 (Homo sapiens)
Gene name (HAGRID: 25)
- HGNC symbol
- HDAC3
- Aliases
- RPD3; HD3; RPD3-2
- Common name
- histone deacetylase 3
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
HDAC3 performs the deacetylation of histones and likely participates in transcriptional regulation [736]. Homologues of HDAC3 have been linked to longevity in yeast [203] and to ageing and caloric restriction in fruit flies [202]. Its relation to human ageing is not known, yet because of its many functions, it is possible that HDAC3 plays some role on ageing.
Cytogenetic information
- Cytogenetic band
- 5q31
- Location
- 141,620,876 bp to 141,636,856 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0001934; positive regulation of protein phosphorylation
GO:0006325; chromatin organization
GO:0006351; transcription, DNA-templated
GO:0006476; protein deacetylation
GO:0007623; circadian rhythm
GO:0010832; negative regulation of myotube differentiation
GO:0031647; regulation of protein stability
GO:0032008; positive regulation of TOR signaling
GO:0042993; positive regulation of transcription factor import into nucleus
GO:0043066; negative regulation of apoptotic process
GO:0044255; cellular lipid metabolic process
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0046329; negative regulation of JNK cascade
GO:0051225; spindle assembly
GO:0070932; histone H3 deacetylation
GO:0071498; cellular response to fluid shear stress
Cellular component: GO:0000118; histone deacetylase complex
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005794; Golgi apparatus
GO:0005829; cytosol
GO:0005876; spindle microtubule
GO:0005886; plasma membrane
GO:0017053; transcriptional repressor complex
Hide GO termsFunction: GO:0003682; chromatin binding
GO:0003714; transcription corepressor activity
GO:0004407; histone deacetylase activity
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0019899; enzyme binding
GO:0030332; cyclin binding
GO:0032041; NAD-dependent histone deacetylase activity (H3-K14 specific)
GO:0033558; protein deacetylase activity
GO:0042826; histone deacetylase binding
GO:0051059; NF-kappaB binding
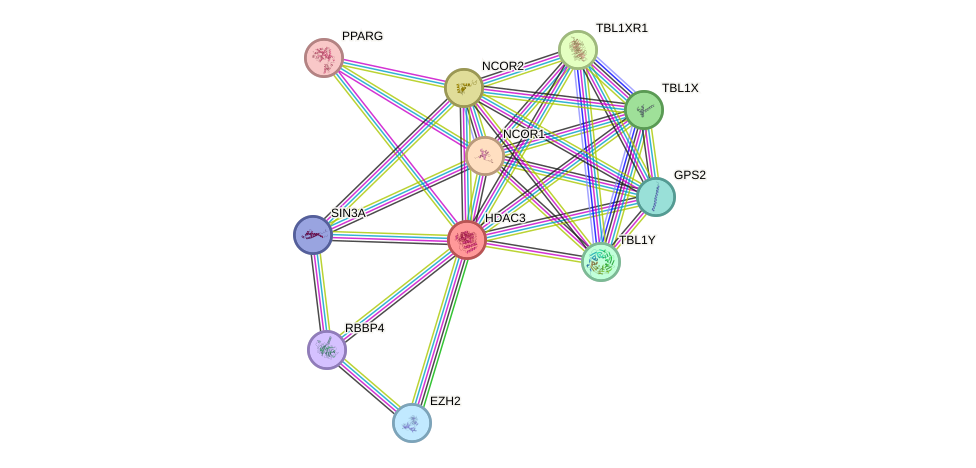
Protein interactions and network
- Protein-protein interacting partners in GenAge
- STAT3, HDAC3, AKT1, MYC, NCOR1, PARP1, PIN1, CREBBP, HIF1A, TOP2B, NFKB1, EP300, PML, PRKDC, AR, RB1, EMD, RELA, MAPK14, SP1, JUN, PIK3R1, APEX1, HSPA8, ESR1, NFKBIA, PPP1CA, PPARG, NCOR2, NFE2L2
- STRING interaction network
Retrieve sequences for HDAC3
Homologs in model organisms
In other databases
- CellAge
- This gene is present as HDAC3