GenAge entry for ABL1 (Homo sapiens)
Gene name (HAGRID: 78)
- HGNC symbol
- ABL1
- Aliases
- JTK7; c-ABL; p150; ABL
- Common name
- ABL proto-oncogene 1, non-receptor tyrosine kinase
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
An important player in cellular proliferation and development, ABL1 also acts as an oncogene [471]. Some results indicate ABL1 might also be involved in DNA repair [474]. Disruption of ABL1 in mice results in severe defects and animals die shortly after birth [472]. Although its exact functions remain elusive, ABL1 has been argued to play a role in ageing due to its interaction with ageing-related genes, such as ATM, and its functions [1292].
Cytogenetic information
- Cytogenetic band
- 9q34.1
- Location
- 130,713,881 bp to 130,887,675 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0001922; B-1 B cell homeostasis
GO:0001934; positive regulation of protein phosphorylation
GO:0002322; B cell proliferation involved in immune response
GO:0002333; transitional one stage B cell differentiation
GO:0006298; mismatch repair
GO:0006355; regulation of transcription, DNA-templated
GO:0006464; cellular protein modification process
GO:0006914; autophagy
GO:0006974; cellular response to DNA damage stimulus
GO:0006975; DNA damage induced protein phosphorylation
GO:0006979; response to oxidative stress
GO:0007050; cell cycle arrest
GO:0007067; mitotic nuclear division
GO:0007173; epidermal growth factor receptor signaling pathway
GO:0007204; positive regulation of cytosolic calcium ion concentration
GO:0008630; intrinsic apoptotic signaling pathway in response to DNA damage
GO:0010506; regulation of autophagy
GO:0018108; peptidyl-tyrosine phosphorylation
GO:0021587; cerebellum morphogenesis
GO:0022408; negative regulation of cell-cell adhesion
GO:0030035; microspike assembly
GO:0030036; actin cytoskeleton organization
GO:0030100; regulation of endocytosis
GO:0030155; regulation of cell adhesion
GO:0030514; negative regulation of BMP signaling pathway
GO:0030516; regulation of axon extension
GO:0031113; regulation of microtubule polymerization
GO:0032956; regulation of actin cytoskeleton organization
GO:0033690; positive regulation of osteoblast proliferation
GO:0034446; substrate adhesion-dependent cell spreading
GO:0034599; cellular response to oxidative stress
GO:0035791; platelet-derived growth factor receptor-beta signaling pathway
GO:0038083; peptidyl-tyrosine autophosphorylation
GO:0038096; Fc-gamma receptor signaling pathway involved in phagocytosis
GO:0042127; regulation of cell proliferation
GO:0042770; signal transduction in response to DNA damage
GO:0043065; positive regulation of apoptotic process
GO:0043123; positive regulation of I-kappaB kinase/NF-kappaB signaling
GO:0043124; negative regulation of I-kappaB kinase/NF-kappaB signaling
GO:0045087; innate immune response
GO:0045184; establishment of protein localization
GO:0045930; negative regulation of mitotic cell cycle
GO:0045931; positive regulation of mitotic cell cycle
GO:0046632; alpha-beta T cell differentiation
GO:0046777; protein autophosphorylation
GO:0048536; spleen development
GO:0048538; thymus development
GO:0048668; collateral sprouting
GO:0050731; positive regulation of peptidyl-tyrosine phosphorylation
GO:0050798; activated T cell proliferation
GO:0050853; B cell receptor signaling pathway
GO:0050885; neuromuscular process controlling balance
GO:0051149; positive regulation of muscle cell differentiation
GO:0051281; positive regulation of release of sequestered calcium ion into cytosol
GO:0051353; positive regulation of oxidoreductase activity
GO:0051444; negative regulation of ubiquitin-protein transferase activity
GO:0051882; mitochondrial depolarization
GO:0060020; Bergmann glial cell differentiation
GO:0070301; cellular response to hydrogen peroxide
GO:0070373; negative regulation of ERK1 and ERK2 cascade
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:0071222; cellular response to lipopolysaccharide
GO:0071901; negative regulation of protein serine/threonine kinase activity
GO:0090135; actin filament branching
GO:1900042; positive regulation of interleukin-2 secretion
GO:1900275; negative regulation of phospholipase C activity
GO:1901216; positive regulation of neuron death
GO:1902715; positive regulation of interferon-gamma secretion
GO:1903053; regulation of extracellular matrix organization
GO:1903351; cellular response to dopamine
GO:1904528; positive regulation of microtubule binding
GO:1904531; positive regulation of actin filament binding
GO:1990051; activation of protein kinase C activity
GO:2000096; positive regulation of Wnt signaling pathway, planar cell polarity pathway
GO:2000145; regulation of cell motility
GO:2000249; regulation of actin cytoskeleton reorganization
GO:2000352; negative regulation of endothelial cell apoptotic process
GO:2000773; negative regulation of cellular senescence
GO:2001020; regulation of response to DNA damage stimulus
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005730; nucleolus
GO:0005737; cytoplasm
GO:0005739; mitochondrion
GO:0005829; cytosol
GO:0015629; actin cytoskeleton
GO:0031234; extrinsic component of cytoplasmic side of plasma membrane
GO:0031252; cell leading edge
GO:0031965; nuclear membrane
GO:0048471; perinuclear region of cytoplasm
Hide GO termsFunction: GO:0000287; magnesium ion binding
GO:0003677; DNA binding
GO:0003785; actin monomer binding
GO:0004515; nicotinate-nucleotide adenylyltransferase activity
GO:0004672; protein kinase activity
GO:0004713; protein tyrosine kinase activity
GO:0004715; non-membrane spanning protein tyrosine kinase activity
GO:0005080; protein kinase C binding
GO:0005102; receptor binding
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0008022; protein C-terminus binding
GO:0017124; SH3 domain binding
GO:0019905; syntaxin binding
GO:0030145; manganese ion binding
GO:0051015; actin filament binding
GO:0051019; mitogen-activated protein kinase binding
GO:0070064; proline-rich region binding
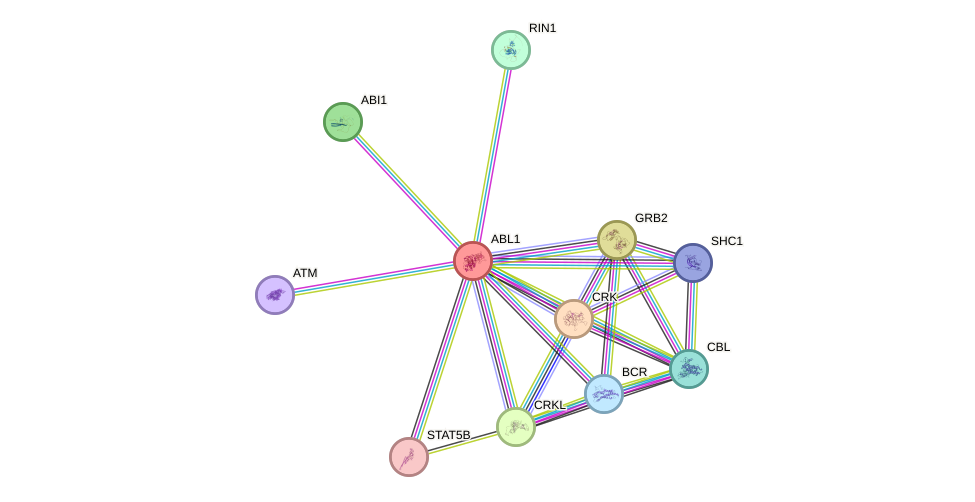
Protein interactions and network
- Protein-protein interacting partners in GenAge
- SHC1, TP53, ATM, EGFR, ERBB2, FOS, PARP1, BRCA1, HSP90AA1, ABL1, RAD51, TERF1, PRKDC, CAT, PCNA, XRCC6, HSPA8, RB1, GRB2, PRDX1, GPX1, HSPD1, YWHAZ, JUN, PIK3R1, CREB1, CDK1, MDM2, NFKBIA, MTOR, CTNNB1, MAPT, ATR, STUB1, TP73
- STRING interaction network
Retrieve sequences for ABL1
Homologs in model organisms
- Caenorhabditis elegans
- abl-1
- Danio rerio
- abl1
- Drosophila melanogaster
- Abl
- Mus musculus
- Abl1
- Rattus norvegicus
- Abl1