GenAge entry for CREB1 (Homo sapiens)
Gene name (HAGRID: 192)
- HGNC symbol
- CREB1
- Aliases
- Common name
- cAMP responsive element binding protein 1
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to the regulation or control of genes previously linked to ageing
- Description
CREB1 is an important transcription factor involved in a variety of processes. CREB1-null mice die shortly after birth [1729]. Results from mice suggest that CREB1 may contribute to type 2 diabetes [841] and neurodegeneration [843]. CREB1 may also regulate some ageing-related genes such as ATM [1013]. Because it is involved in so many different functions, a role for CREB1 in human ageing is plausible though so far unverified.
Cytogenetic information
- Cytogenetic band
- 2q34
- Location
- 207,529,892 bp to 207,605,560 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0001666; response to hypoxia
GO:0006366; transcription from RNA polymerase II promoter
GO:0006468; protein phosphorylation
GO:0007165; signal transduction
GO:0007179; transforming growth factor beta receptor signaling pathway
GO:0007411; axon guidance
GO:0007568; aging
GO:0007595; lactation
GO:0007613; memory
GO:0007623; circadian rhythm
GO:0008361; regulation of cell size
GO:0008542; visual learning
GO:0010033; response to organic substance
GO:0010944; negative regulation of transcription by competitive promoter binding
GO:0014823; response to activity
GO:0016032; viral process
GO:0021983; pituitary gland development
GO:0032916; positive regulation of transforming growth factor beta3 production
GO:0033363; secretory granule organization
GO:0033762; response to glucagon
GO:0034670; chemotaxis to arachidonic acid
GO:0035094; response to nicotine
GO:0035729; cellular response to hepatocyte growth factor stimulus
GO:0036120; cellular response to platelet-derived growth factor stimulus
GO:0040018; positive regulation of multicellular organism growth
GO:0042493; response to drug
GO:0042752; regulation of circadian rhythm
GO:0043065; positive regulation of apoptotic process
GO:0045600; positive regulation of fat cell differentiation
GO:0045672; positive regulation of osteoclast differentiation
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045899; positive regulation of RNA polymerase II transcriptional preinitiation complex assembly
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0046887; positive regulation of hormone secretion
GO:0046889; positive regulation of lipid biosynthetic process
GO:0048145; regulation of fibroblast proliferation
GO:0050821; protein stabilization
GO:0055025; positive regulation of cardiac muscle tissue development
GO:0060251; regulation of glial cell proliferation
GO:0060430; lung saccule development
GO:0060509; Type I pneumocyte differentiation
GO:0071294; cellular response to zinc ion
GO:0071398; cellular response to fatty acid
GO:1900273; positive regulation of long-term synaptic potentiation
GO:1901215; negative regulation of neuron death
GO:1902065; response to L-glutamate
GO:1990090; cellular response to nerve growth factor stimulus
GO:1990314; cellular response to insulin-like growth factor stimulus
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005719; nuclear euchromatin
GO:0005759; mitochondrial matrix
GO:0030424; axon
GO:1990589; ATF4-CREB1 transcription factor complex
Hide GO termsFunction: GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0000980; RNA polymerase II distal enhancer sequence-specific DNA binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001102; RNA polymerase II activating transcription factor binding
GO:0001190; transcriptional activator activity, RNA polymerase II transcription factor binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0003705; transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding
GO:0003712; transcription cofactor activity
GO:0005515; protein binding
GO:0019899; enzyme binding
GO:0030544; Hsp70 protein binding
GO:0035035; histone acetyltransferase binding
GO:0035497; cAMP response element binding
GO:0042802; identical protein binding
GO:1990763; arrestin family protein binding
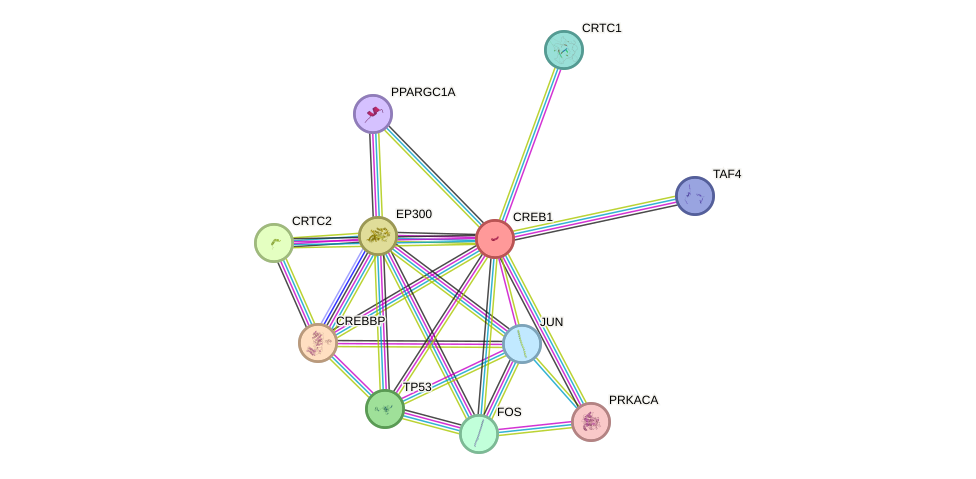
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, ATM, AKT1, NCOR1, JUND, BRCA1, CREBBP, NR3C1, ABL1, UBE2I, CEBPB, EP300, GSK3B, HTT, SIRT1, HDAC1, MAPK14, JUN, CREB1, ATF2, HDAC2, CTNNB1, PPP1CA, ATR, GSK3A
- STRING interaction network