GenAge entry for MAPK14 (Homo sapiens)
Gene name (HAGRID: 168)
- HGNC symbol
- MAPK14
- Aliases
- PRKM14; p38; Mxi2; PRKM15; CSPB1; CSBP1; CSBP2
- Common name
- mitogen-activated protein kinase 14
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence directly linking the gene product to ageing in a cellular model system
- Description
Also known as p38, MAPK14 is activated in response to cellular stress and it can phosphorylate a number of transcription factors. It is also involved in the production of IL6 [137], and is an important player in cellular senescence [140]. In-vitro inhibition of p38 enhances the proliferative potential of mice muscle stem cells and restores the age-declined regenerative potential of stem cells populations [3640]. In senescent CD8+ T cells its inhibition increases proliferation, telomerase activity, mitochondrial biogenesis, and fitness, in a mTOR-independent manner [3641]. Mice without MAPK14 are anaemic due to failed definitive erythropoiesis [773]. Transgenic mice with induced MAPK14 activity in the heart died in 7-9 weeks [775]. MAPK14's responsiveness may change with ageing in mouse cells [176]. A role for MAPK14 in human ageing is so far unknown.
Cytogenetic information
- Cytogenetic band
- 6p21
- Location
- 36,027,677 bp to 36,111,236 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000077; DNA damage checkpoint
GO:0000187; activation of MAPK activity
GO:0000902; cell morphogenesis
GO:0001502; cartilage condensation
GO:0001525; angiogenesis
GO:0001890; placenta development
GO:0002062; chondrocyte differentiation
GO:0006006; glucose metabolic process
GO:0006351; transcription, DNA-templated
GO:0006357; regulation of transcription from RNA polymerase II promoter
GO:0006915; apoptotic process
GO:0006928; movement of cell or subcellular component
GO:0006935; chemotaxis
GO:0007165; signal transduction
GO:0007166; cell surface receptor signaling pathway
GO:0007178; transmembrane receptor protein serine/threonine kinase signaling pathway
GO:0007265; Ras protein signal transduction
GO:0007519; skeletal muscle tissue development
GO:0010628; positive regulation of gene expression
GO:0010831; positive regulation of myotube differentiation
GO:0014835; myoblast differentiation involved in skeletal muscle regeneration
GO:0018105; peptidyl-serine phosphorylation
GO:0019395; fatty acid oxidation
GO:0030278; regulation of ossification
GO:0030316; osteoclast differentiation
GO:0031281; positive regulation of cyclase activity
GO:0031663; lipopolysaccharide-mediated signaling pathway
GO:0032495; response to muramyl dipeptide
GO:0035556; intracellular signal transduction
GO:0035924; cellular response to vascular endothelial growth factor stimulus
GO:0035994; response to muscle stretch
GO:0038066; p38MAPK cascade
GO:0042307; positive regulation of protein import into nucleus
GO:0042770; signal transduction in response to DNA damage
GO:0043536; positive regulation of blood vessel endothelial cell migration
GO:0045648; positive regulation of erythrocyte differentiation
GO:0045663; positive regulation of myoblast differentiation
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0046326; positive regulation of glucose import
GO:0048010; vascular endothelial growth factor receptor signaling pathway
GO:0051090; regulation of sequence-specific DNA binding transcription factor activity
GO:0051146; striated muscle cell differentiation
GO:0051149; positive regulation of muscle cell differentiation
GO:0060045; positive regulation of cardiac muscle cell proliferation
GO:0070935; 3'-UTR-mediated mRNA stabilization
GO:0071222; cellular response to lipopolysaccharide
GO:0071479; cellular response to ionizing radiation
GO:0090090; negative regulation of canonical Wnt signaling pathway
GO:0090336; positive regulation of brown fat cell differentiation
GO:0090400; stress-induced premature senescence
GO:0098586; cellular response to virus
GO:1900015; regulation of cytokine production involved in inflammatory response
GO:1901741; positive regulation of myoblast fusion
GO:1901796; regulation of signal transduction by p53 class mediator
GO:2000379; positive regulation of reactive oxygen species metabolic process
GO:2001184; positive regulation of interleukin-12 secretion
Cellular component: GO:0000922; spindle pole
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005739; mitochondrion
GO:0005829; cytosol
GO:0070062; extracellular exosome
Hide GO termsFunction: GO:0004674; protein serine/threonine kinase activity
GO:0004707; MAP kinase activity
GO:0004708; MAP kinase kinase activity
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0019899; enzyme binding
GO:0019903; protein phosphatase binding
GO:0051525; NFAT protein binding
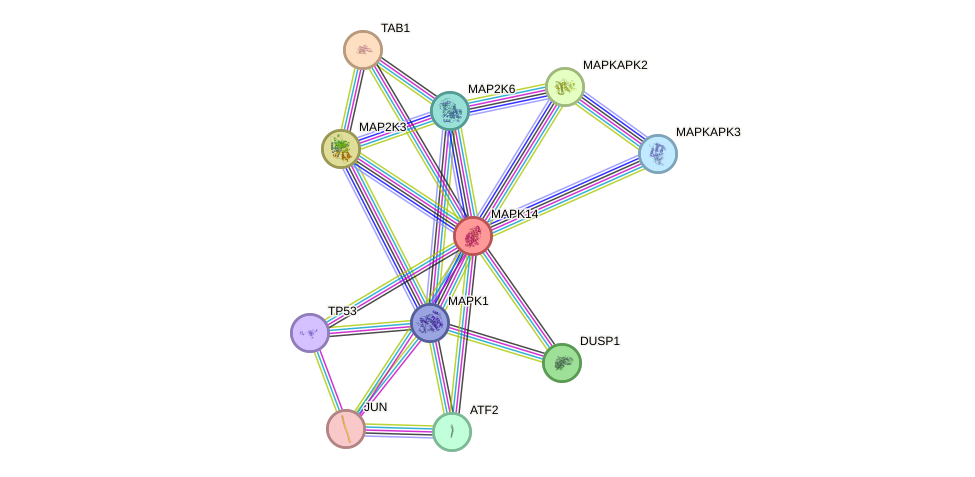
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, ATM, PTPN11, HDAC3, AKT1, EGFR, EP300, EEF2, RB1, RECQL4, RELA, HDAC1, HSPD1, MAPK14, SP1, JUN, MAPK3, BMI1, EEF1A1, CREB1, ATF2, DDIT3, MAX, NFKBIA, HTRA2, IKBKB, SQSTM1, RICTOR
- STRING interaction network
Retrieve sequences for MAPK14
Homologs in model organisms
In other databases
- CellAge
- This gene is present as MAPK14