GenAge entry for PDGFRA (Homo sapiens)
Gene name (HAGRID: 285)
- HGNC symbol
- PDGFRA
- Aliases
- CD140a; PDGFR2
- Common name
- platelet-derived growth factor receptor, alpha polypeptide
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
The PDGFRA gene encodes a cell surface tyrosine kinase receptor for members of the platelet-derived growth factor family. These growth factors are mitogens for cells of mesenchymal origin. PDGFRA binds PDGFB and thus may be related to age-related changes in the heart [363]. With age, there is a decline in the PDGFRA levels of beta-cell in mice, accompanied by a reduction in beta-cell replication. The conditional inactivation of Pdgfra accelerates these changes, preventing neonatal beta-cell expansion and adult beta-cell regeneration. Similar results were observed in human beta-cells [2224]. Receptor tyrosine kinases have been found to be expressed in metastatic colon cancer [2225] and PDGFRA mutations were found in gastrointestinal tumours [2226]. PDGFRA has not been directly related to ageing, however.
Cytogenetic information
- Cytogenetic band
- 4q12
- Location
- 54,229,097 bp to 54,298,245 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000165; MAPK cascade
GO:0001553; luteinization
GO:0001701; in utero embryonic development
GO:0001775; cell activation
GO:0002244; hematopoietic progenitor cell differentiation
GO:0007204; positive regulation of cytosolic calcium ion concentration
GO:0008210; estrogen metabolic process
GO:0008284; positive regulation of cell proliferation
GO:0010544; negative regulation of platelet activation
GO:0010863; positive regulation of phospholipase C activity
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0014068; positive regulation of phosphatidylinositol 3-kinase signaling
GO:0016032; viral process
GO:0018108; peptidyl-tyrosine phosphorylation
GO:0023019; signal transduction involved in regulation of gene expression
GO:0030198; extracellular matrix organization
GO:0030324; lung development
GO:0030325; adrenal gland development
GO:0030335; positive regulation of cell migration
GO:0030539; male genitalia development
GO:0033327; Leydig cell differentiation
GO:0034614; cellular response to reactive oxygen species
GO:0035790; platelet-derived growth factor receptor-alpha signaling pathway
GO:0038091; positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway
GO:0042060; wound healing
GO:0042475; odontogenesis of dentin-containing tooth
GO:0043547; positive regulation of GTPase activity
GO:0043552; positive regulation of phosphatidylinositol 3-kinase activity
GO:0045740; positive regulation of DNA replication
GO:0046777; protein autophosphorylation
GO:0046854; phosphatidylinositol phosphorylation
GO:0048008; platelet-derived growth factor receptor signaling pathway
GO:0048015; phosphatidylinositol-mediated signaling
GO:0048146; positive regulation of fibroblast proliferation
GO:0048557; embryonic digestive tract morphogenesis
GO:0048701; embryonic cranial skeleton morphogenesis
GO:0048704; embryonic skeletal system morphogenesis
GO:0050920; regulation of chemotaxis
GO:0055003; cardiac myofibril assembly
GO:0060021; palate development
GO:0060325; face morphogenesis
GO:0060326; cell chemotaxis
GO:0061298; retina vasculature development in camera-type eye
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:0070527; platelet aggregation
GO:0071230; cellular response to amino acid stimulus
GO:0072277; metanephric glomerular capillary formation
GO:2000249; regulation of actin cytoskeleton reorganization
GO:2000739; regulation of mesenchymal stem cell differentiation
Cellular component: GO:0005634; nucleus
GO:0005737; cytoplasm
GO:0005886; plasma membrane
GO:0005887; integral component of plasma membrane
GO:0005902; microvillus
GO:0009897; external side of plasma membrane
GO:0016020; membrane
GO:0031226; intrinsic component of plasma membrane
GO:0043234; protein complex
Hide GO termsFunction: GO:0004672; protein kinase activity
GO:0004714; transmembrane receptor protein tyrosine kinase activity
GO:0005018; platelet-derived growth factor alpha-receptor activity
GO:0005021; vascular endothelial growth factor-activated receptor activity
GO:0005088; Ras guanyl-nucleotide exchange factor activity
GO:0005161; platelet-derived growth factor receptor binding
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0032403; protein complex binding
GO:0038085; vascular endothelial growth factor binding
GO:0042803; protein homodimerization activity
GO:0046934; phosphatidylinositol-4,5-bisphosphate 3-kinase activity
GO:0048407; platelet-derived growth factor binding
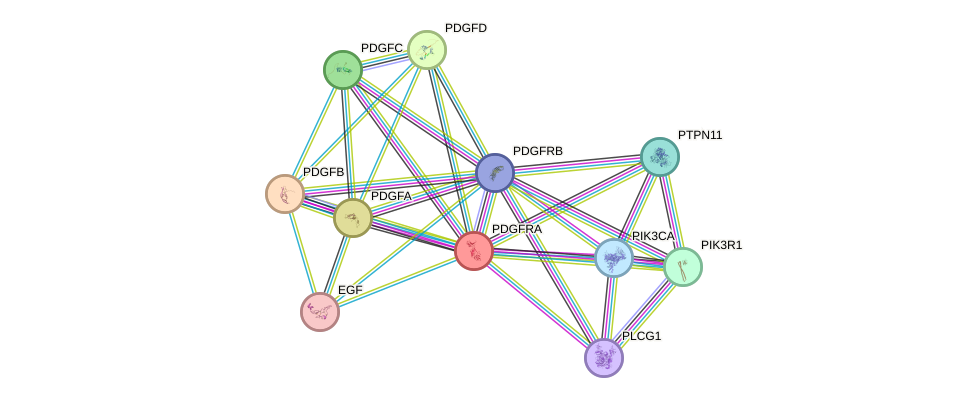
Protein interactions and network
- Protein-protein interacting partners in GenAge
- STAT3, EGFR, PDGFB, PDGFRB, PTEN, GRB2, PIK3R1, CLU, PDGFRA
- STRING interaction network