GenAge entry for PTGS2 (Homo sapiens)
Gene name (HAGRID: 198)
- HGNC symbol
- PTGS2
- Aliases
- COX2
- Common name
- prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on indirect or inconclusive evidence linking the gene product to ageing in humans or in one or more model systems
- Description
PTGS2 may play a role in inflammation. PTGS2-null mice developed severe nephropathy [864] and female reproductive failures [1734]. Age-related changes in PTGS2 have been reported in various tissues in mice [863]. PTGS2 has not been directly related to human ageing.
Cytogenetic information
- Cytogenetic band
- 1q25.2-q25
- Location
- 186,671,812 bp to 186,680,427 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0001516; prostaglandin biosynthetic process
GO:0001525; angiogenesis
GO:0006693; prostaglandin metabolic process
GO:0006769; nicotinamide metabolic process
GO:0006928; movement of cell or subcellular component
GO:0006954; inflammatory response
GO:0006979; response to oxidative stress
GO:0007566; embryo implantation
GO:0007612; learning
GO:0007613; memory
GO:0008217; regulation of blood pressure
GO:0008285; negative regulation of cell proliferation
GO:0009750; response to fructose
GO:0010042; response to manganese ion
GO:0010226; response to lithium ion
GO:0010575; positive regulation of vascular endothelial growth factor production
GO:0019233; sensory perception of pain
GO:0019371; cyclooxygenase pathway
GO:0019372; lipoxygenase pathway
GO:0030282; bone mineralization
GO:0030728; ovulation
GO:0031394; positive regulation of prostaglandin biosynthetic process
GO:0031622; positive regulation of fever generation
GO:0031915; positive regulation of synaptic plasticity
GO:0032227; negative regulation of synaptic transmission, dopaminergic
GO:0032355; response to estradiol
GO:0032496; response to lipopolysaccharide
GO:0033280; response to vitamin D
GO:0034612; response to tumor necrosis factor
GO:0034644; cellular response to UV
GO:0035633; maintenance of permeability of blood-brain barrier
GO:0042346; positive regulation of NF-kappaB import into nucleus
GO:0042493; response to drug
GO:0042633; hair cycle
GO:0043065; positive regulation of apoptotic process
GO:0045429; positive regulation of nitric oxide biosynthetic process
GO:0045786; negative regulation of cell cycle
GO:0045907; positive regulation of vasoconstriction
GO:0045986; negative regulation of smooth muscle contraction
GO:0045987; positive regulation of smooth muscle contraction
GO:0046697; decidualization
GO:0048661; positive regulation of smooth muscle cell proliferation
GO:0050727; regulation of inflammatory response
GO:0050873; brown fat cell differentiation
GO:0051384; response to glucocorticoid
GO:0051926; negative regulation of calcium ion transport
GO:0051968; positive regulation of synaptic transmission, glutamatergic
GO:0055114; oxidation-reduction process
GO:0070542; response to fatty acid
GO:0071260; cellular response to mechanical stimulus
GO:0071318; cellular response to ATP
GO:0071456; cellular response to hypoxia
GO:0071498; cellular response to fluid shear stress
GO:0071636; positive regulation of transforming growth factor beta production
GO:0090050; positive regulation of cell migration involved in sprouting angiogenesis
GO:0090271; positive regulation of fibroblast growth factor production
GO:0090336; positive regulation of brown fat cell differentiation
GO:0090362; positive regulation of platelet-derived growth factor production
GO:0098869; cellular oxidant detoxification
Cellular component: GO:0005634; nucleus
GO:0005737; cytoplasm
GO:0005783; endoplasmic reticulum
GO:0005788; endoplasmic reticulum lumen
GO:0005789; endoplasmic reticulum membrane
GO:0005901; caveola
GO:0031090; organelle membrane
GO:0043005; neuron projection
GO:0043234; protein complex
Hide GO termsFunction: GO:0004601; peroxidase activity
GO:0004666; prostaglandin-endoperoxide synthase activity
GO:0005515; protein binding
GO:0008289; lipid binding
GO:0019899; enzyme binding
GO:0020037; heme binding
GO:0042803; protein homodimerization activity
GO:0046872; metal ion binding
GO:0050473; arachidonate 15-lipoxygenase activity
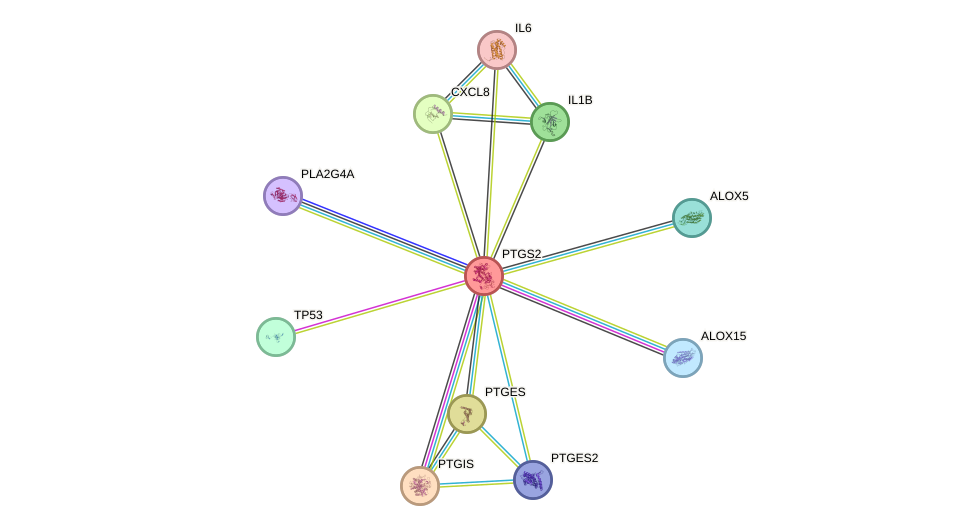
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, VCP, EP300, APP, MAPK3, PTGS2, SUMO1, CTNNB1
- STRING interaction network
Retrieve sequences for PTGS2
Homologs in model organisms
In other databases
- CellAge
- This gene is present as PTGS2