GenAge entry for RET (Homo sapiens)
Gene name (HAGRID: 56)
- HGNC symbol
- RET
- Aliases
- PTC; CDHF12; RET51; CDHR16; HSCR1; MEN2A; MTC1; MEN2B
- Common name
- ret proto-oncogene
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
A receptor tyrosine kinase that is also considered an oncogene, RET promotes cellular growth. Its levels increase with age in the nervous system [215], and it has been related to cancer, but at present there is no evidence to suggest it plays a major role in human ageing.
Cytogenetic information
- Cytogenetic band
- 10q11.2
- Location
- 43,077,069 bp to 43,130,349 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000165; MAPK cascade
GO:0001657; ureteric bud development
GO:0001755; neural crest cell migration
GO:0001838; embryonic epithelial tube formation
GO:0006468; protein phosphorylation
GO:0007156; homophilic cell adhesion via plasma membrane adhesion molecules
GO:0007158; neuron cell-cell adhesion
GO:0007165; signal transduction
GO:0007169; transmembrane receptor protein tyrosine kinase signaling pathway
GO:0007497; posterior midgut development
GO:0010976; positive regulation of neuron projection development
GO:0014042; positive regulation of neuron maturation
GO:0018108; peptidyl-tyrosine phosphorylation
GO:0030155; regulation of cell adhesion
GO:0030335; positive regulation of cell migration
GO:0033619; membrane protein proteolysis
GO:0033630; positive regulation of cell adhesion mediated by integrin
GO:0035799; ureter maturation
GO:0042493; response to drug
GO:0042551; neuron maturation
GO:0043547; positive regulation of GTPase activity
GO:0045793; positive regulation of cell size
GO:0045893; positive regulation of transcription, DNA-templated
GO:0048265; response to pain
GO:0048484; enteric nervous system development
GO:0050770; regulation of axonogenesis
GO:0060041; retina development in camera-type eye
GO:0060384; innervation
GO:0061146; Peyer's patch morphogenesis
GO:0071300; cellular response to retinoic acid
GO:0072300; positive regulation of metanephric glomerulus development
GO:0097021; lymphocyte migration into lymphoid organs
GO:1903263; positive regulation of serine phosphorylation of STAT3 protein
GO:2001241; positive regulation of extrinsic apoptotic signaling pathway in absence of ligand
Cellular component: GO:0005737; cytoplasm
GO:0005769; early endosome
GO:0005886; plasma membrane
GO:0005887; integral component of plasma membrane
GO:0010008; endosome membrane
GO:0030424; axon
GO:0030425; dendrite
GO:0043025; neuronal cell body
GO:0043231; intracellular membrane-bounded organelle
GO:0043235; receptor complex
GO:0045121; membrane raft
Hide GO termsFunction: GO:0004713; protein tyrosine kinase activity
GO:0004714; transmembrane receptor protein tyrosine kinase activity
GO:0004872; receptor activity
GO:0005088; Ras guanyl-nucleotide exchange factor activity
GO:0005509; calcium ion binding
GO:0005515; protein binding
GO:0005524; ATP binding
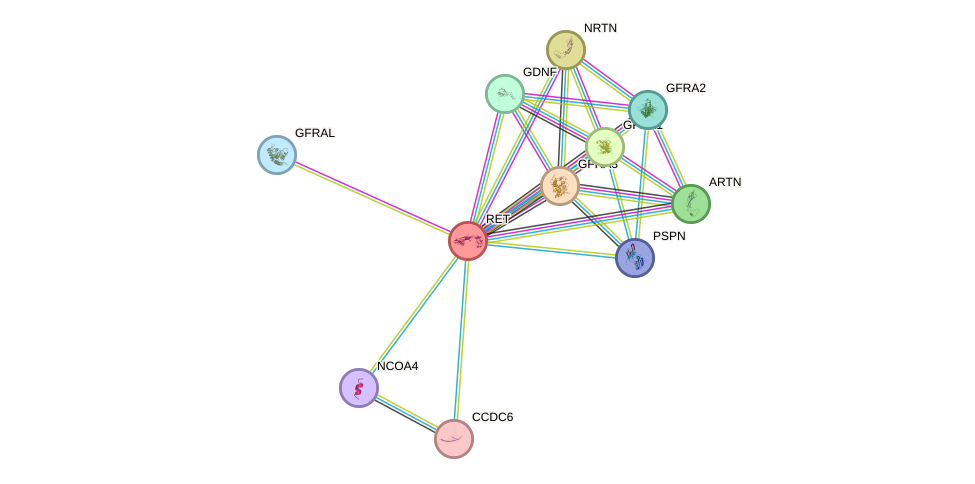
Protein interactions and network
- Protein-protein interacting partners in GenAge
- SHC1, STAT3, EGFR, RET, HSP90AA1, GRB2, PTK2, MAPK3
- STRING interaction network