A Conserved Molecular Signature of Caloric Restriction
Caloric restriction (CR) is the only non-genetic intervention known to delay aging in a wide range of model organism and has triggered the development of CR mimetics by the pharmaceutical industry. We performed a meta-analysis of CR microarray studies in mammals to obtain a comprehensive picture of CR-induced changes and identify the most promising genes and functional categories for mediating the life-extending effects of CR. Our results reveal a complex array of CR-induced changes and highlight key processes associated with CR, such as growth hormone signalling, lipid metabolism and immune response. Moreover, our results highlight novel associations with CR, such as retinol metabolism and copper ion detoxification. Analyses of our signatures by integrating co-expression data, information on genetic mutants, and transcription factor binding site analysis reveals candidate regulators of transcriptional modules in CR. Overall, our conserved molecular signatures of CR provide a comprehensive picture of CR-induced changes and help understand its regulatory mechanisms.
In this page you will find the results derived from our meta-analysis, including the supplementary material, previously described in:
Plank, M., et al. (2012) A meta-analysis of caloric restriction gene expression profiles to infer common signatures and regulatory mechanisms. Molecular BioSystems 8(4):1339-1349.
Searchable database:
The significant genes from this work are also available as a searchable database. As incidated in the paper, mouse gene symbols and names were used.
Supplementary material:
mbs12_supplementary.zip, zipped file with the meta-profiling results for genes and GO categories, data sources, additional details regarding the methodology, etc.
Flow diagram of our meta-analysis method
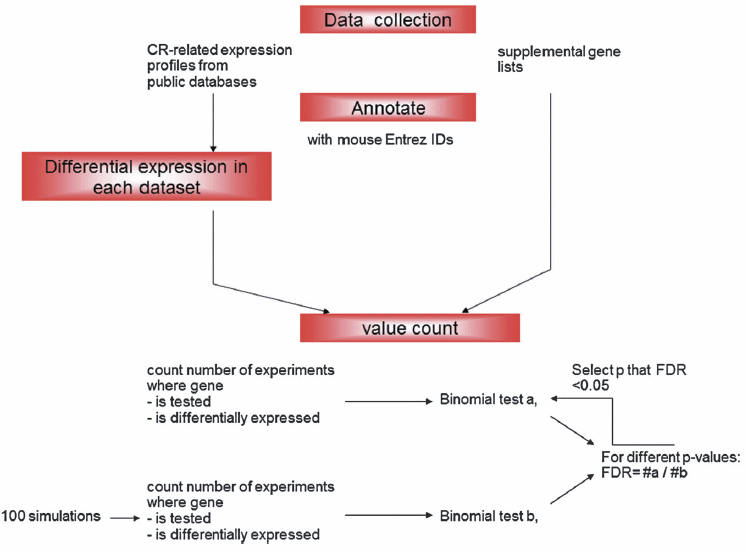
Please refer to the Methods section of the paper for additional details.