A Conserved Gene Expression Signature of Mammalian Aging
Gene expression changes likely play important roles in aging and could serve as biomarkers of physiological decline and disease, yet age-related gene expression profiles tend to be noisy, making it difficult to identify key genes and processes. In this work, we sought to address this problem by performing a meta-analysis of 27 age-related gene expression profiles from mice, rats, and humans. By analyzing the combined profiles from these studies we were able to identify common molecular signatures of aging: genes consistently over- or underexpressed with age in multiple mammalian tissues and species. We also characterized the biological processes associated with these signatures and we found that age-related gene expression changes most notably involve an overexpression of genes related to inflammation and the lysosome as well as an underexpression of genes related to mitochondrial energy metabolism. Overall, our work reveals previously unknown transcriptional changes with age and helps paint a better picture of how changes in gene expression relate to the process of aging.
In this page you will find the results derived from our meta-analysis, including the supplementary material, previously described in:
de Magalhaes, J. P., Curado, J., Church, G. M. (2009) Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25:875-881.
Searchable database:
The significant genes from this work are also available as a searchable database. As incidated in the paper human gene symbols and names were used (where possible).
Supplementary material:
signatures_supplement.zip, zipped file with the meta-profiling results for genes and GO categories plus additional details regarding the methodology.
Flow diagram of our meta-analysis method
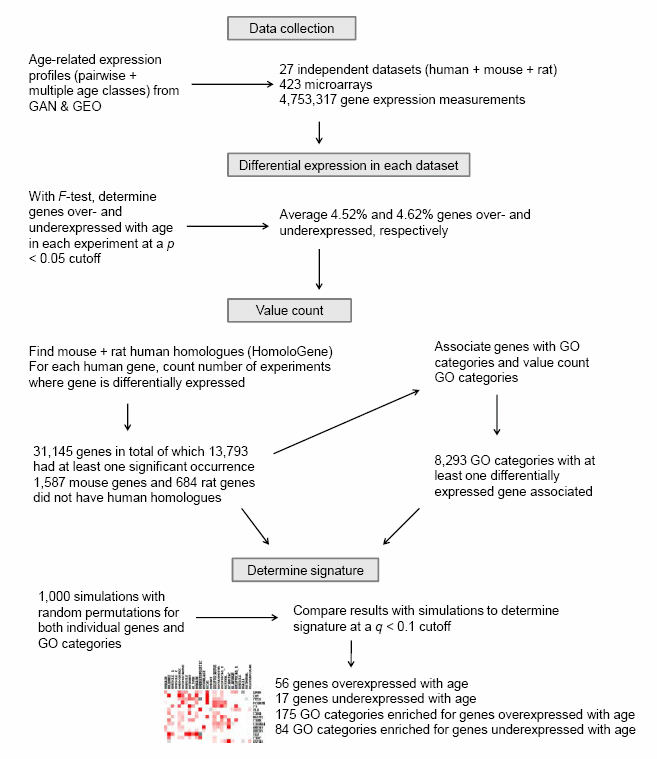
Please refer to the Methods section of the paper for additional details.