GenAge entry for ESR1 (Homo sapiens)
Gene name (HAGRID: 216)
- HGNC symbol
- ESR1
- Aliases
- NR3A1; Era; ESR
- Common name
- estrogen receptor 1
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to the regulation or control of genes previously linked to ageing
- Description
ESR1 is a transcription factor that mediates the actions of estrogen. Mice without ESR1 were infertile and showed a variety of changes [1193]. Polymorphisms in the human ESR1 gene have been associated with cardiovascular disease [1740]. ESR1 has also been found to be upregulated in Alzheimer's disease [1185]. A role for ESR1 in human ageing is plausible though at present largely obscure.
Cytogenetic information
- Cytogenetic band
- 6q25.1
- Location
- 151,807,678 bp to 152,103,273 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0001547; antral ovarian follicle growth
GO:0002064; epithelial cell development
GO:0006338; chromatin remodeling
GO:0006351; transcription, DNA-templated
GO:0006355; regulation of transcription, DNA-templated
GO:0006366; transcription from RNA polymerase II promoter
GO:0006367; transcription initiation from RNA polymerase II promoter
GO:0007165; signal transduction
GO:0007200; phospholipase C-activating G-protein coupled receptor signaling pathway
GO:0007204; positive regulation of cytosolic calcium ion concentration
GO:0008209; androgen metabolic process
GO:0008584; male gonad development
GO:0010629; negative regulation of gene expression
GO:0010863; positive regulation of phospholipase C activity
GO:0030518; intracellular steroid hormone receptor signaling pathway
GO:0030520; intracellular estrogen receptor signaling pathway
GO:0032355; response to estradiol
GO:0034121; regulation of toll-like receptor signaling pathway
GO:0042981; regulation of apoptotic process
GO:0043124; negative regulation of I-kappaB kinase/NF-kappaB signaling
GO:0043433; negative regulation of sequence-specific DNA binding transcription factor activity
GO:0043627; response to estrogen
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0048146; positive regulation of fibroblast proliferation
GO:0050727; regulation of inflammatory response
GO:0051091; positive regulation of sequence-specific DNA binding transcription factor activity
GO:0060065; uterus development
GO:0060068; vagina development
GO:0060523; prostate epithelial cord elongation
GO:0060527; prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis
GO:0060687; regulation of branching involved in prostate gland morphogenesis
GO:0060745; mammary gland branching involved in pregnancy
GO:0060749; mammary gland alveolus development
GO:0060750; epithelial cell proliferation involved in mammary gland duct elongation
GO:0071168; protein localization to chromatin
GO:0071391; cellular response to estrogen stimulus
GO:0071392; cellular response to estradiol stimulus
GO:1903799; negative regulation of production of miRNAs involved in gene silencing by miRNA
Cellular component: GO:0000790; nuclear chromatin
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005794; Golgi apparatus
GO:0005886; plasma membrane
GO:0016020; membrane
GO:0016021; integral component of membrane
GO:0035327; transcriptionally active chromatin
Hide GO termsFunction: GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0001046; core promoter sequence-specific DNA binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0003682; chromatin binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0003707; steroid hormone receptor activity
GO:0005496; steroid binding
GO:0005515; protein binding
GO:0008013; beta-catenin binding
GO:0008134; transcription factor binding
GO:0008270; zinc ion binding
GO:0019899; enzyme binding
GO:0030235; nitric-oxide synthase regulator activity
GO:0030284; estrogen receptor activity
GO:0034056; estrogen response element binding
GO:0038052; RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding
GO:0042802; identical protein binding
GO:0051117; ATPase binding
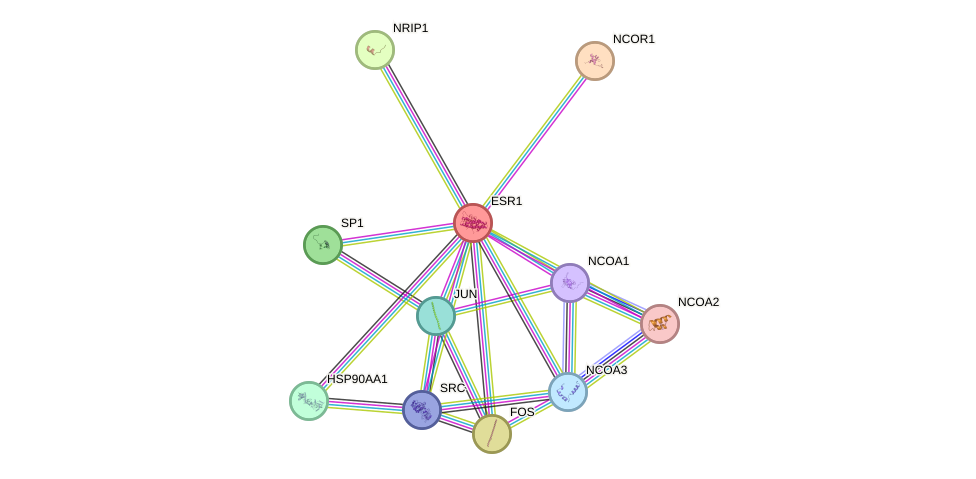
Protein interactions and network
- Protein-protein interacting partners in GenAge
- SHC1, TP53, IGF1R, STAT3, STAT5A, IRS1, IRS2, AKT1, MYC, EGFR, ERBB2, FOS, HDAC3, PARP1, BRCA1, PTEN, CREBBP, HIF1A, NCOR1, HSP90AA1, TOP2A, TOP2B, TOP1, RAD51, UBE2I, EP300, EEF2, PRKDC, CAT, XRCC5, XRCC6, HSPA8, EMD, FOXO1, AIFM1, SIRT1, HDAC1, HSPD1, SP1, JUN, MED1, FOXO4, PIK3R1, TBP, HBP1, SIN3A, E2F1, HDAC2, MDM2, RELA, VCP, H2AFX, CEBPA, CEBPB, ESR1, CTNNB1, DBN1, PPP1CA, STUB1, PPARGC1A, NCOR2, CDKN1A, PIK3CA, CDK7
- STRING interaction network