GenAge entry for MAPK3 (Homo sapiens)
Gene name (HAGRID: 175)
- HGNC symbol
- MAPK3
- Aliases
- ERK1; p44mapk; p44erk1; PRKM3
- Common name
- mitogen-activated protein kinase 3
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
Also called ERK1, MAPK3 is involved in stress response signalling and maybe in cell cycle control. MAPK3-null mice are viable and fertile but have defective thymocyte maturation [788]. Changes in stress response have been reported during murine ageing that could be a result of changes in MAPK3 [346]. The relevance of MAPK3 to human ageing remains undetermined.
Cytogenetic information
- Cytogenetic band
- 16p11.2
- Location
- 30,114,105 bp to 30,123,309 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000165; MAPK cascade
GO:0000187; activation of MAPK activity
GO:0000189; MAPK import into nucleus
GO:0001934; positive regulation of protein phosphorylation
GO:0006361; transcription initiation from RNA polymerase I promoter
GO:0006461; protein complex assembly
GO:0006468; protein phosphorylation
GO:0006915; apoptotic process
GO:0006975; DNA damage induced protein phosphorylation
GO:0007049; cell cycle
GO:0007411; axon guidance
GO:0008543; fibroblast growth factor receptor signaling pathway
GO:0009636; response to toxic substance
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0016032; viral process
GO:0016310; phosphorylation
GO:0018105; peptidyl-serine phosphorylation
GO:0019233; sensory perception of pain
GO:0019369; arachidonic acid metabolic process
GO:0030168; platelet activation
GO:0030278; regulation of ossification
GO:0030509; BMP signaling pathway
GO:0030878; thyroid gland development
GO:0031281; positive regulation of cyclase activity
GO:0031663; lipopolysaccharide-mediated signaling pathway
GO:0032212; positive regulation of telomere maintenance via telomerase
GO:0032872; regulation of stress-activated MAPK cascade
GO:0033129; positive regulation of histone phosphorylation
GO:0035066; positive regulation of histone acetylation
GO:0038083; peptidyl-tyrosine autophosphorylation
GO:0038095; Fc-epsilon receptor signaling pathway
GO:0038096; Fc-gamma receptor signaling pathway involved in phagocytosis
GO:0042473; outer ear morphogenesis
GO:0043330; response to exogenous dsRNA
GO:0045727; positive regulation of translation
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0048538; thymus development
GO:0051090; regulation of sequence-specific DNA binding transcription factor activity
GO:0051216; cartilage development
GO:0051493; regulation of cytoskeleton organization
GO:0051973; positive regulation of telomerase activity
GO:0060020; Bergmann glial cell differentiation
GO:0060324; face development
GO:0060397; JAK-STAT cascade involved in growth hormone signaling pathway
GO:0060425; lung morphogenesis
GO:0060440; trachea formation
GO:0061308; cardiac neural crest cell development involved in heart development
GO:0070371; ERK1 and ERK2 cascade
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:0070498; interleukin-1-mediated signaling pathway
GO:0070849; response to epidermal growth factor
GO:0071260; cellular response to mechanical stimulus
GO:0072584; caveolin-mediated endocytosis
GO:0090170; regulation of Golgi inheritance
GO:1900034; regulation of cellular response to heat
GO:1904355; positive regulation of telomere capping
GO:1904417; positive regulation of xenophagy
GO:2000641; regulation of early endosome to late endosome transport
GO:2000657; negative regulation of apolipoprotein binding
Cellular component: GO:0005634; nucleus
GO:0005635; nuclear envelope
GO:0005654; nucleoplasm
GO:0005739; mitochondrion
GO:0005769; early endosome
GO:0005770; late endosome
GO:0005794; Golgi apparatus
GO:0005829; cytosol
GO:0005856; cytoskeleton
GO:0005901; caveola
GO:0005925; focal adhesion
GO:0015630; microtubule cytoskeleton
GO:0031143; pseudopodium
GO:0043234; protein complex
GO:0070062; extracellular exosome
Hide GO termsFunction: GO:0001784; phosphotyrosine binding
GO:0004674; protein serine/threonine kinase activity
GO:0004707; MAP kinase activity
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0016301; kinase activity
GO:0019902; phosphatase binding
GO:0097110; scaffold protein binding
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, MYC, INSR, RET, BRCA1, CREBBP, VCP, PML, HTT, SNCG, MAPK14, SP1, JUN, MAPK3, ATF2, PTGS2, SIRT6, CHEK2, TP53BP1, HTRA2
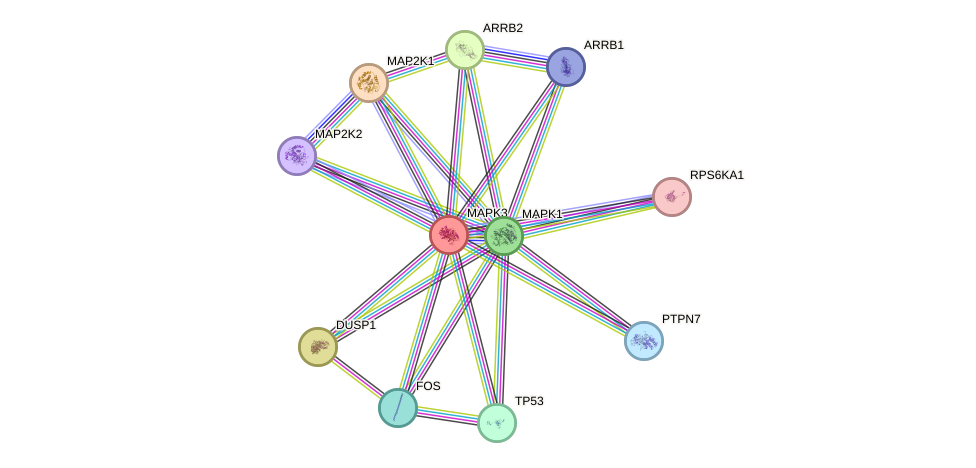
- STRING interaction network
Retrieve sequences for MAPK3
Homologs in model organisms
- Caenorhabditis elegans
- mpk-1
- Drosophila melanogaster
- rl
- Mus musculus
- Mapk3
- Rattus norvegicus
- Mapk3
- Saccharomyces cerevisiae
- KSS1
- Schizosaccharomyces pombe
- spk1
In other databases
- GenAge model organism genes
- LongevityMap
- This gene is present as MAPK3