GenAge entry for PDGFB (Homo sapiens)
Gene name (HAGRID: 47)
- HGNC symbol
- PDGFB
- Aliases
- SSV; SIS
- Common name
- platelet-derived growth factor beta polypeptide
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to the regulation or control of genes previously linked to ageing
- Description
PDGFB is a mitogen and oncogene involved in cell survival, development, and healing [363]. Results from rats suggest it may be related to age-related changes in the heart [362], and PDGF's anti-apoptotic signalling involves several pathways and genes that in turn have been associated with ageing [333]. PDGFB's exact relevance to human ageing, however, is unknown.
Cytogenetic information
- Cytogenetic band
- 22q13.1
- Location
- 39,223,680 bp to 39,244,952 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000165; MAPK cascade
GO:0001892; embryonic placenta development
GO:0001938; positive regulation of endothelial cell proliferation
GO:0002548; monocyte chemotaxis
GO:0002576; platelet degranulation
GO:0003104; positive regulation of glomerular filtration
GO:0006468; protein phosphorylation
GO:0007179; transforming growth factor beta receptor signaling pathway
GO:0007507; heart development
GO:0008284; positive regulation of cell proliferation
GO:0009611; response to wounding
GO:0010512; negative regulation of phosphatidylinositol biosynthetic process
GO:0010544; negative regulation of platelet activation
GO:0010628; positive regulation of gene expression
GO:0010629; negative regulation of gene expression
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0014068; positive regulation of phosphatidylinositol 3-kinase signaling
GO:0014911; positive regulation of smooth muscle cell migration
GO:0018105; peptidyl-serine phosphorylation
GO:0018108; peptidyl-tyrosine phosphorylation
GO:0030097; hemopoiesis
GO:0030198; extracellular matrix organization
GO:0030335; positive regulation of cell migration
GO:0031954; positive regulation of protein autophosphorylation
GO:0032091; negative regulation of protein binding
GO:0032147; activation of protein kinase activity
GO:0032148; activation of protein kinase B activity
GO:0035655; interleukin-18-mediated signaling pathway
GO:0035793; positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway
GO:0038001; paracrine signaling
GO:0043406; positive regulation of MAP kinase activity
GO:0043410; positive regulation of MAPK cascade
GO:0043536; positive regulation of blood vessel endothelial cell migration
GO:0043547; positive regulation of GTPase activity
GO:0043552; positive regulation of phosphatidylinositol 3-kinase activity
GO:0045737; positive regulation of cyclin-dependent protein serine/threonine kinase activity
GO:0045740; positive regulation of DNA replication
GO:0045840; positive regulation of mitotic nuclear division
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045893; positive regulation of transcription, DNA-templated
GO:0046854; phosphatidylinositol phosphorylation
GO:0048008; platelet-derived growth factor receptor signaling pathway
GO:0048015; phosphatidylinositol-mediated signaling
GO:0048146; positive regulation of fibroblast proliferation
GO:0048661; positive regulation of smooth muscle cell proliferation
GO:0050731; positive regulation of peptidyl-tyrosine phosphorylation
GO:0050918; positive chemotaxis
GO:0050921; positive regulation of chemotaxis
GO:0051781; positive regulation of cell division
GO:0060326; cell chemotaxis
GO:0061098; positive regulation of protein tyrosine kinase activity
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:0070528; protein kinase C signaling
GO:0071363; cellular response to growth factor stimulus
GO:0071506; cellular response to mycophenolic acid
GO:0072126; positive regulation of glomerular mesangial cell proliferation
GO:0072255; metanephric glomerular mesangial cell development
GO:0072593; reactive oxygen species metabolic process
GO:0090280; positive regulation of calcium ion import
GO:1900127; positive regulation of hyaluronan biosynthetic process
GO:1902894; negative regulation of pri-miRNA transcription from RNA polymerase II promoter
GO:1902895; positive regulation of pri-miRNA transcription from RNA polymerase II promoter
GO:1904707; positive regulation of vascular smooth muscle cell proliferation
GO:1904754; positive regulation of vascular associated smooth muscle cell migration
GO:1905064; negative regulation of vascular smooth muscle cell differentiation
GO:1905176; positive regulation of vascular smooth muscle cell dedifferentiation
GO:2000379; positive regulation of reactive oxygen species metabolic process
GO:2000573; positive regulation of DNA biosynthetic process
GO:2000591; positive regulation of metanephric mesenchymal cell migration
Cellular component: GO:0000139; Golgi membrane
GO:0005576; extracellular region
GO:0005615; extracellular space
GO:0005737; cytoplasm
GO:0005788; endoplasmic reticulum lumen
GO:0009986; cell surface
GO:0016323; basolateral plasma membrane
GO:0031012; extracellular matrix
GO:0031093; platelet alpha granule lumen
Hide GO termsFunction: GO:0005088; Ras guanyl-nucleotide exchange factor activity
GO:0005161; platelet-derived growth factor receptor binding
GO:0005515; protein binding
GO:0005518; collagen binding
GO:0008083; growth factor activity
GO:0016176; superoxide-generating NADPH oxidase activator activity
GO:0042056; chemoattractant activity
GO:0042802; identical protein binding
GO:0042803; protein homodimerization activity
GO:0046934; phosphatidylinositol-4,5-bisphosphate 3-kinase activity
GO:0046982; protein heterodimerization activity
GO:0048407; platelet-derived growth factor binding
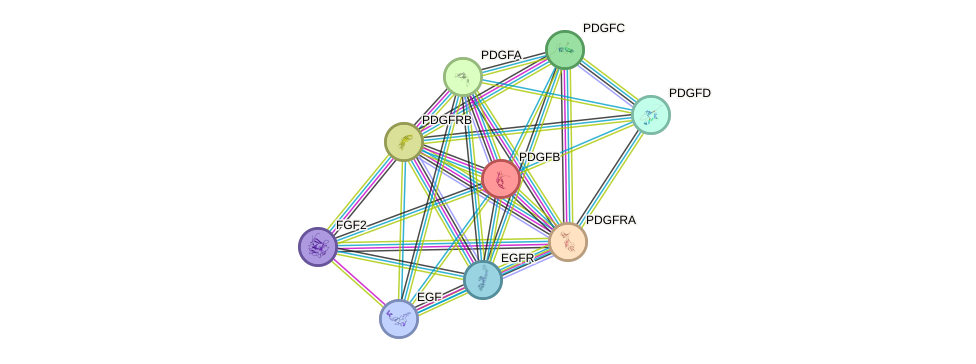
Protein interactions and network
Retrieve sequences for PDGFB
Homologs in model organisms
In other databases
- CellAge
- This gene is present as PDGFB