GenAge entry for ATF2 (Homo sapiens)
Gene name (HAGRID: 193)
- HGNC symbol
- ATF2
- Aliases
- TREB7; CRE-BP1; HB16; CREB2
- Common name
- activating transcription factor 2
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
ATF2 is an important transcription factor involved in a variety of functions that may also be involved in oxidative stress response [135] and cellular growth arrest and senescence [137]. It has not been directly related to human ageing.
Cytogenetic information
- Cytogenetic band
- 2q32
- Location
- 175,072,250 bp to 175,168,206 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0003151; outflow tract morphogenesis
GO:0006355; regulation of transcription, DNA-templated
GO:0006357; regulation of transcription from RNA polymerase II promoter
GO:0006366; transcription from RNA polymerase II promoter
GO:0006970; response to osmotic stress
GO:0006974; cellular response to DNA damage stimulus
GO:0009414; response to water deprivation
GO:0016573; histone acetylation
GO:0031573; intra-S DNA damage checkpoint
GO:0032915; positive regulation of transforming growth factor beta2 production
GO:0043525; positive regulation of neuron apoptotic process
GO:0045444; fat cell differentiation
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0050680; negative regulation of epithelial cell proliferation
GO:0051090; regulation of sequence-specific DNA binding transcription factor activity
GO:0051091; positive regulation of sequence-specific DNA binding transcription factor activity
GO:0060612; adipose tissue development
GO:0097186; amelogenesis
GO:1902110; positive regulation of mitochondrial membrane permeability involved in apoptotic process
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005741; mitochondrial outer membrane
GO:0035861; site of double-strand break
Hide GO termsFunction: GO:0000977; RNA polymerase II regulatory region sequence-specific DNA binding
GO:0000980; RNA polymerase II distal enhancer sequence-specific DNA binding
GO:0001076; transcription factor activity, RNA polymerase II transcription factor binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001102; RNA polymerase II activating transcription factor binding
GO:0001158; enhancer sequence-specific DNA binding
GO:0001228; transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding
GO:0003682; chromatin binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0003705; transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding
GO:0003713; transcription coactivator activity
GO:0004402; histone acetyltransferase activity
GO:0005515; protein binding
GO:0008140; cAMP response element binding protein binding
GO:0019901; protein kinase binding
GO:0035497; cAMP response element binding
GO:0046872; metal ion binding
GO:0046982; protein heterodimerization activity
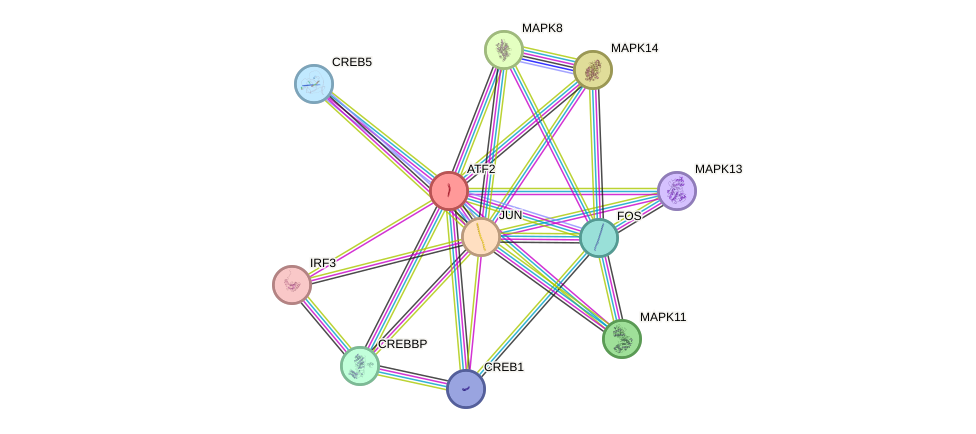
Protein interactions and network
- Protein-protein interacting partners in GenAge
- ATM, PTPN1, NBN, FOS, CREBBP, HSP90AA1, UBE2I, CEBPA, CEBPB, EP300, PML, EEF2, PCNA, RB1, APP, ATP5O, GSTP1, HSPD1, MAPK8, MAPK14, JUN, MAPK9, MAPK3, CREB1, ATF2, DDIT3, SUMO1, H2AFX, CTNNB1, MLH1
- STRING interaction network