GenAge entry for EGF (Homo sapiens)
Gene name (HAGRID: 48)
- HGNC symbol
- EGF
- Aliases
- Common name
- epidermal growth factor
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to the regulation or control of genes previously linked to ageing
- Description
EGF stimulates the growth of several epidermal and epithelial cell types. It is also involved in differentiation [366]. Expression of EGF in transgenic mice causes stunted growth [367]. An attenuation of EGF signalling has been reported with age [346], but it is unclear whether these are causes or effects of ageing.
Cytogenetic information
- Cytogenetic band
- 4q25
- Location
- 109,912,884 bp to 110,012,962 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000165; MAPK cascade
GO:0000186; activation of MAPKK activity
GO:0000187; activation of MAPK activity
GO:0001525; angiogenesis
GO:0002092; positive regulation of receptor internalization
GO:0002576; platelet degranulation
GO:0006260; DNA replication
GO:0007165; signal transduction
GO:0007171; activation of transmembrane receptor protein tyrosine kinase activity
GO:0007173; epidermal growth factor receptor signaling pathway
GO:0007262; STAT protein import into nucleus
GO:0008284; positive regulation of cell proliferation
GO:0010800; positive regulation of peptidyl-threonine phosphorylation
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0018108; peptidyl-tyrosine phosphorylation
GO:0021940; positive regulation of cerebellar granule cell precursor proliferation
GO:0035413; positive regulation of catenin import into nucleus
GO:0038128; ERBB2 signaling pathway
GO:0042059; negative regulation of epidermal growth factor receptor signaling pathway
GO:0042327; positive regulation of phosphorylation
GO:0043388; positive regulation of DNA binding
GO:0043406; positive regulation of MAP kinase activity
GO:0043547; positive regulation of GTPase activity
GO:0044332; Wnt signaling pathway involved in dorsal/ventral axis specification
GO:0045741; positive regulation of epidermal growth factor-activated receptor activity
GO:0045840; positive regulation of mitotic nuclear division
GO:0045893; positive regulation of transcription, DNA-templated
GO:0046854; phosphatidylinositol phosphorylation
GO:0048015; phosphatidylinositol-mediated signaling
GO:0048754; branching morphogenesis of an epithelial tube
GO:0051048; negative regulation of secretion
GO:0060070; canonical Wnt signaling pathway
GO:0060749; mammary gland alveolus development
GO:0070371; ERK1 and ERK2 cascade
GO:0090279; regulation of calcium ion import
GO:0090370; negative regulation of cholesterol efflux
GO:1900127; positive regulation of hyaluronan biosynthetic process
GO:2000008; regulation of protein localization to cell surface
GO:2000060; positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process
GO:2000145; regulation of cell motility
Cellular component: GO:0005576; extracellular region
GO:0005615; extracellular space
GO:0005765; lysosomal membrane
GO:0005886; plasma membrane
GO:0016021; integral component of membrane
GO:0031093; platelet alpha granule lumen
GO:0043235; receptor complex
GO:0070062; extracellular exosome
Hide GO termsFunction: GO:0004713; protein tyrosine kinase activity
GO:0005088; Ras guanyl-nucleotide exchange factor activity
GO:0005154; epidermal growth factor receptor binding
GO:0005509; calcium ion binding
GO:0005515; protein binding
GO:0008083; growth factor activity
GO:0017147; Wnt-protein binding
GO:0030297; transmembrane receptor protein tyrosine kinase activator activity
GO:0042813; Wnt-activated receptor activity
GO:0046934; phosphatidylinositol-4,5-bisphosphate 3-kinase activity
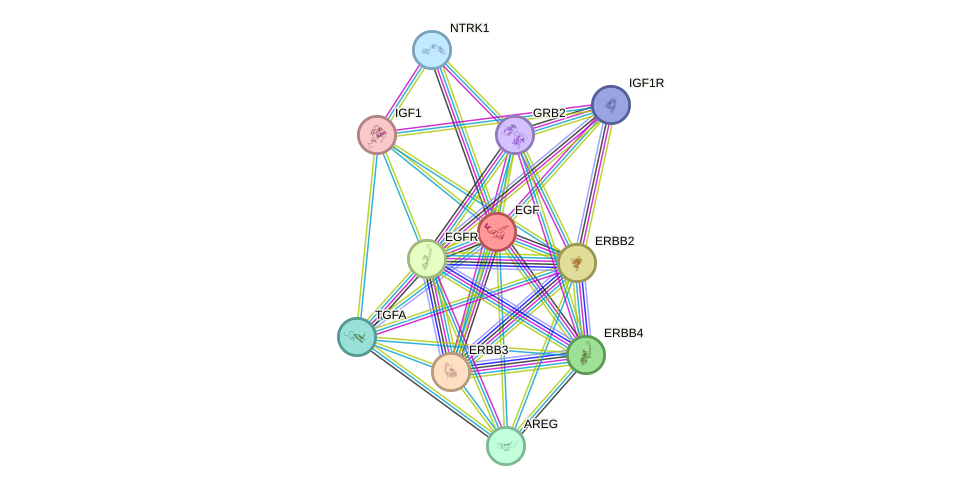
Protein interactions and network
- Protein-protein interacting partners in GenAge
- LMNA, EGFR, ERBB2, EGF, UBE2I, EMD, LMNB1
- STRING interaction network
Retrieve sequences for EGF
Homologs in model organisms
In other databases
- LongevityMap
- This gene is present as EGF