GenAge entry for FGFR1 (Homo sapiens)
Gene name (HAGRID: 169)
- HGNC symbol
- FGFR1
- Aliases
- H2; H3; H4; H5; CEK; FLG; BFGFR; N-SAM; CD331; FLT2; KAL2
- Common name
- fibroblast growth factor receptor 1
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on indirect or inconclusive evidence linking the gene product to ageing in humans or in one or more model systems
- Description
FGFR1 is the receptor of the fibroblast growth factor. Mice with an attenuation of FGFR1 signalling develop diabetes with age and exhibit a decreased number of beta-cells [777]. Lower levels of FGFR1 in mice have also been related to craniofacial defects [778]. Mutations in the human FGFR1 gene have been associated with Pfeiffer [780] and Kallmann syndromes [779]. While FGFR1 could play a role in some age-related diseases, such as diabetes, its involvement in human ageing remains to be determined.
Cytogenetic information
- Cytogenetic band
- 8p12
- Location
- 38,411,138 bp to 38,468,834 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0000165; MAPK cascade
GO:0001501; skeletal system development
GO:0001525; angiogenesis
GO:0001657; ureteric bud development
GO:0001701; in utero embryonic development
GO:0001759; organ induction
GO:0001764; neuron migration
GO:0002053; positive regulation of mesenchymal cell proliferation
GO:0002062; chondrocyte differentiation
GO:0006351; transcription, DNA-templated
GO:0006468; protein phosphorylation
GO:0007037; vacuolar phosphate transport
GO:0007605; sensory perception of sound
GO:0008284; positive regulation of cell proliferation
GO:0008543; fibroblast growth factor receptor signaling pathway
GO:0010518; positive regulation of phospholipase activity
GO:0010863; positive regulation of phospholipase C activity
GO:0010976; positive regulation of neuron projection development
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0014068; positive regulation of phosphatidylinositol 3-kinase signaling
GO:0016477; cell migration
GO:0018108; peptidyl-tyrosine phosphorylation
GO:0021847; ventricular zone neuroblast division
GO:0030326; embryonic limb morphogenesis
GO:0030901; midbrain development
GO:0035607; fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development
GO:0036092; phosphatidylinositol-3-phosphate biosynthetic process
GO:0042472; inner ear morphogenesis
GO:0042473; outer ear morphogenesis
GO:0042474; middle ear morphogenesis
GO:0043009; chordate embryonic development
GO:0043406; positive regulation of MAP kinase activity
GO:0043410; positive regulation of MAPK cascade
GO:0043547; positive regulation of GTPase activity
GO:0045595; regulation of cell differentiation
GO:0045666; positive regulation of neuron differentiation
GO:0046777; protein autophosphorylation
GO:0046854; phosphatidylinositol phosphorylation
GO:0048015; phosphatidylinositol-mediated signaling
GO:0048339; paraxial mesoderm development
GO:0048378; regulation of lateral mesodermal cell fate specification
GO:0048469; cell maturation
GO:0048705; skeletal system morphogenesis
GO:0048762; mesenchymal cell differentiation
GO:0060045; positive regulation of cardiac muscle cell proliferation
GO:0060117; auditory receptor cell development
GO:0060445; branching involved in salivary gland morphogenesis
GO:0060484; lung-associated mesenchyme development
GO:0060665; regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling
GO:0070640; vitamin D3 metabolic process
GO:0090080; positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway
GO:0090272; negative regulation of fibroblast growth factor production
GO:1903465; positive regulation of mitotic cell cycle DNA replication
GO:2000546; positive regulation of endothelial cell chemotaxis to fibroblast growth factor
GO:2000830; positive regulation of parathyroid hormone secretion
GO:2001239; regulation of extrinsic apoptotic signaling pathway in absence of ligand
Cellular component: GO:0005576; extracellular region
GO:0005634; nucleus
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0005887; integral component of plasma membrane
GO:0016021; integral component of membrane
GO:0031410; cytoplasmic vesicle
GO:0043235; receptor complex
Hide GO termsFunction: GO:0004713; protein tyrosine kinase activity
GO:0005007; fibroblast growth factor-activated receptor activity
GO:0005088; Ras guanyl-nucleotide exchange factor activity
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0008201; heparin binding
GO:0016303; 1-phosphatidylinositol-3-kinase activity
GO:0017134; fibroblast growth factor binding
GO:0042802; identical protein binding
GO:0042803; protein homodimerization activity
GO:0046934; phosphatidylinositol-4,5-bisphosphate 3-kinase activity
GO:0090722; receptor-receptor interaction
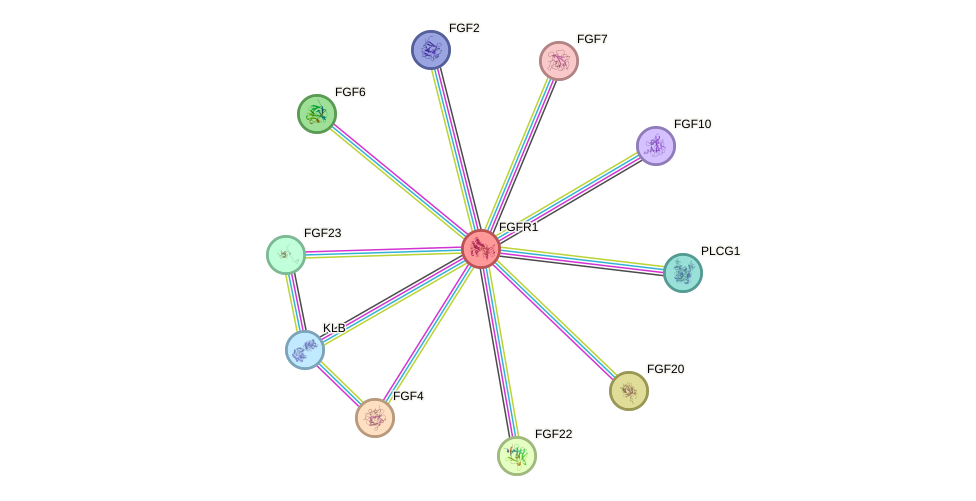
Protein interactions and network
- Protein-protein interacting partners in GenAge
- STAT3, CREBBP, HSP90AA1, FGFR1, PIK3R1, JAK2, CTNNB1, FGF23
- STRING interaction network