GenAge entry for HRAS (Homo sapiens)
Gene name (HAGRID: 38)
- HGNC symbol
- HRAS
- Aliases
- HRAS1
- Common name
- Harvey rat sarcoma viral oncogene homolog
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence directly linking the gene product to ageing in a non-mammalian animal model
- Description
HRAS is a member of the RAS family of oncogenes and probably plays a role in cell growth and differentiation. In roundworms, the RAS pathway has been shown to influence development and ageing in conjunction with INS/IGF1 [1517], and RAS signalling has been associated with brain degeneration in fruit flies [345]. Although HRAS is involved in cancer and cellular senescence [339], its role in human ageing is unknown.
Cytogenetic information
- Cytogenetic band
- 11p15.5
- Location
- 532,242 bp to 535,550 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000165; MAPK cascade
GO:0001934; positive regulation of protein phosphorylation
GO:0002223; stimulatory C-type lectin receptor signaling pathway
GO:0006897; endocytosis
GO:0006935; chemotaxis
GO:0007050; cell cycle arrest
GO:0007093; mitotic cell cycle checkpoint
GO:0007165; signal transduction
GO:0007166; cell surface receptor signaling pathway
GO:0007173; epidermal growth factor receptor signaling pathway
GO:0007265; Ras protein signal transduction
GO:0007411; axon guidance
GO:0008283; cell proliferation
GO:0008284; positive regulation of cell proliferation
GO:0008285; negative regulation of cell proliferation
GO:0009887; animal organ morphogenesis
GO:0010629; negative regulation of gene expression
GO:0030335; positive regulation of cell migration
GO:0032729; positive regulation of interferon-gamma production
GO:0034260; negative regulation of GTPase activity
GO:0035900; response to isolation stress
GO:0038095; Fc-epsilon receptor signaling pathway
GO:0038128; ERBB2 signaling pathway
GO:0042088; T-helper 1 type immune response
GO:0042832; defense response to protozoan
GO:0043406; positive regulation of MAP kinase activity
GO:0043410; positive regulation of MAPK cascade
GO:0043524; negative regulation of neuron apoptotic process
GO:0043547; positive regulation of GTPase activity
GO:0045740; positive regulation of DNA replication
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0046330; positive regulation of JNK cascade
GO:0046579; positive regulation of Ras protein signal transduction
GO:0048013; ephrin receptor signaling pathway
GO:0048169; regulation of long-term neuronal synaptic plasticity
GO:0050679; positive regulation of epithelial cell proliferation
GO:0050852; T cell receptor signaling pathway
GO:0050900; leukocyte migration
GO:0051291; protein heterooligomerization
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:0090303; positive regulation of wound healing
GO:0090398; cellular senescence
GO:0097193; intrinsic apoptotic signaling pathway
GO:1900029; positive regulation of ruffle assembly
GO:2000251; positive regulation of actin cytoskeleton reorganization
GO:2000630; positive regulation of miRNA metabolic process
Cellular component: GO:0000139; Golgi membrane
GO:0005634; nucleus
GO:0005737; cytoplasm
GO:0005794; Golgi apparatus
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0048471; perinuclear region of cytoplasm
Hide GO termsFunction: GO:0003924; GTPase activity
GO:0005515; protein binding
GO:0005525; GTP binding
GO:0008022; protein C-terminus binding
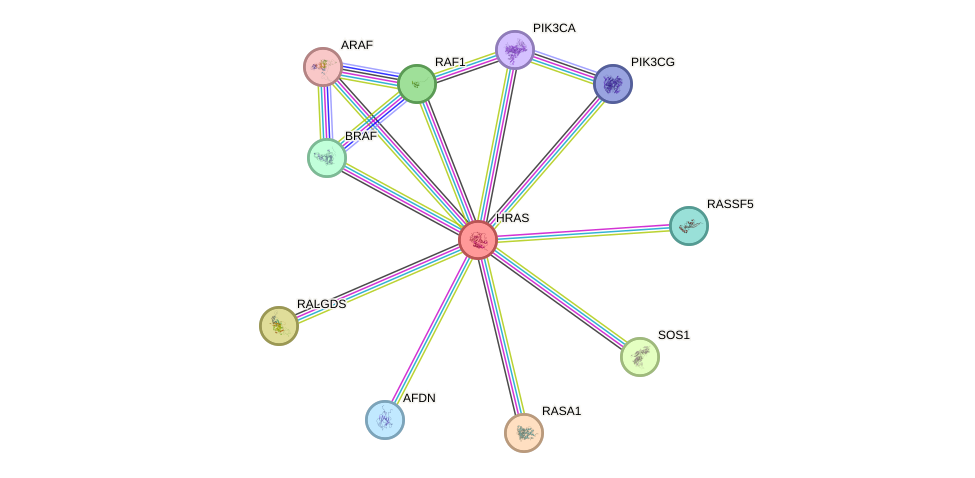
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, PLAU, INSR, MAPK8, PIK3R1, PIK3CA
- STRING interaction network