GenAge entry for NBN (Homo sapiens)
Gene name (HAGRID: 44)
- HGNC symbol
- NBN
- Aliases
- ATV; AT-V2; AT-V1; NBS; NBS1
- Common name
- nibrin
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
Mutations in NBN cause Nijmegen breakage syndrome, a chromosomal instability syndrome that has been suggested as progeroid [55]. NBN is involved in DNA repair, probably of double-strand breaks, in conjunction with other proteins associated with ageing [894]. While not proven, NBN could potentially play some role in human ageing.
Cytogenetic information
- Cytogenetic band
- 8q21
- Location
- 89,933,336 bp to 89,984,671 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000077; DNA damage checkpoint
GO:0000723; telomere maintenance
GO:0000724; double-strand break repair via homologous recombination
GO:0000729; DNA double-strand break processing
GO:0000731; DNA synthesis involved in DNA repair
GO:0000732; strand displacement
GO:0001832; blastocyst growth
GO:0006260; DNA replication
GO:0006302; double-strand break repair
GO:0006303; double-strand break repair via nonhomologous end joining
GO:0007050; cell cycle arrest
GO:0007093; mitotic cell cycle checkpoint
GO:0007095; mitotic G2 DNA damage checkpoint
GO:0008283; cell proliferation
GO:0030174; regulation of DNA-dependent DNA replication initiation
GO:0030330; DNA damage response, signal transduction by p53 class mediator
GO:0031860; telomeric 3' overhang formation
GO:0031954; positive regulation of protein autophosphorylation
GO:0032206; positive regulation of telomere maintenance
GO:0032508; DNA duplex unwinding
GO:0033674; positive regulation of kinase activity
GO:0045190; isotype switching
GO:0050885; neuromuscular process controlling balance
GO:0051321; meiotic cell cycle
GO:0097193; intrinsic apoptotic signaling pathway
GO:1901796; regulation of signal transduction by p53 class mediator
GO:1904354; negative regulation of telomere capping
Cellular component: GO:0000784; nuclear chromosome, telomeric region
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005657; replication fork
GO:0005730; nucleolus
GO:0005829; cytosol
GO:0016605; PML body
GO:0030870; Mre11 complex
GO:0035861; site of double-strand break
GO:0042405; nuclear inclusion body
Hide GO termsFunction: GO:0003684; damaged DNA binding
GO:0004003; ATP-dependent DNA helicase activity
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0047485; protein N-terminus binding
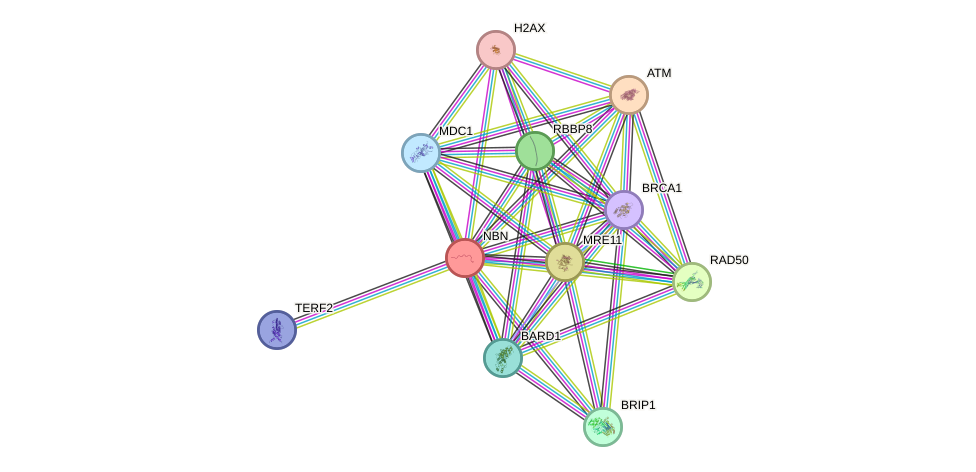
Protein interactions and network
- Protein-protein interacting partners in GenAge
- ATM, WRN, BRCA1, RPA1, BLM, EP300, TERF1, PRKDC, XRCC5, TERF2, SIRT1, BMI1, ATF2, MDM2, H2AFX, MTOR, ATR, MLH1, RICTOR
- STRING interaction network