GenAge entry for PDGFRB (Homo sapiens)
Gene name (HAGRID: 51)
- HGNC symbol
- PDGFRB
- Aliases
- JTK12; CD140b; PDGFR1; PDGFR
- Common name
- platelet-derived growth factor receptor, beta polypeptide
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
PDGFRB binds PDGFB and thus may be related to age-related changes in the heart [363]. PDGFRB is also an important player in development [379].
Cytogenetic information
- Cytogenetic band
- 5q33.1
- Location
- 150,113,839 bp to 150,155,859 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000165; MAPK cascade
GO:0006024; glycosaminoglycan biosynthetic process
GO:0007165; signal transduction
GO:0007186; G-protein coupled receptor signaling pathway
GO:0007568; aging
GO:0008284; positive regulation of cell proliferation
GO:0008584; male gonad development
GO:0009636; response to toxic substance
GO:0010863; positive regulation of phospholipase C activity
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0014068; positive regulation of phosphatidylinositol 3-kinase signaling
GO:0014911; positive regulation of smooth muscle cell migration
GO:0016477; cell migration
GO:0018108; peptidyl-tyrosine phosphorylation
GO:0030335; positive regulation of cell migration
GO:0032355; response to estradiol
GO:0032516; positive regulation of phosphoprotein phosphatase activity
GO:0032526; response to retinoic acid
GO:0032956; regulation of actin cytoskeleton organization
GO:0032967; positive regulation of collagen biosynthetic process
GO:0034405; response to fluid shear stress
GO:0035025; positive regulation of Rho protein signal transduction
GO:0035441; cell migration involved in vasculogenesis
GO:0035789; metanephric mesenchymal cell migration
GO:0035791; platelet-derived growth factor receptor-beta signaling pathway
GO:0035793; positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway
GO:0035909; aorta morphogenesis
GO:0036120; cellular response to platelet-derived growth factor stimulus
GO:0038091; positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway
GO:0042060; wound healing
GO:0042542; response to hydrogen peroxide
GO:0043065; positive regulation of apoptotic process
GO:0043066; negative regulation of apoptotic process
GO:0043406; positive regulation of MAP kinase activity
GO:0043547; positive regulation of GTPase activity
GO:0043552; positive regulation of phosphatidylinositol 3-kinase activity
GO:0043627; response to estrogen
GO:0045840; positive regulation of mitotic nuclear division
GO:0046488; phosphatidylinositol metabolic process
GO:0046777; protein autophosphorylation
GO:0046854; phosphatidylinositol phosphorylation
GO:0048008; platelet-derived growth factor receptor signaling pathway
GO:0048015; phosphatidylinositol-mediated signaling
GO:0048146; positive regulation of fibroblast proliferation
GO:0048661; positive regulation of smooth muscle cell proliferation
GO:0048839; inner ear development
GO:0050921; positive regulation of chemotaxis
GO:0055003; cardiac myofibril assembly
GO:0055093; response to hyperoxia
GO:0060326; cell chemotaxis
GO:0060437; lung growth
GO:0060981; cell migration involved in coronary angiogenesis
GO:0061298; retina vasculature development in camera-type eye
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:0071670; smooth muscle cell chemotaxis
GO:0072075; metanephric mesenchyme development
GO:0072262; metanephric glomerular mesangial cell proliferation involved in metanephros development
GO:0072277; metanephric glomerular capillary formation
GO:0072278; metanephric comma-shaped body morphogenesis
GO:0072284; metanephric S-shaped body morphogenesis
GO:0090280; positive regulation of calcium ion import
GO:2000379; positive regulation of reactive oxygen species metabolic process
GO:2000491; positive regulation of hepatic stellate cell activation
GO:2000573; positive regulation of DNA biosynthetic process
Cellular component: GO:0005634; nucleus
GO:0005737; cytoplasm
GO:0005886; plasma membrane
GO:0005925; focal adhesion
GO:0009986; cell surface
GO:0016020; membrane
GO:0016021; integral component of membrane
GO:0016324; apical plasma membrane
GO:0031226; intrinsic component of plasma membrane
GO:0031410; cytoplasmic vesicle
GO:0043202; lysosomal lumen
GO:0070062; extracellular exosome
Hide GO termsFunction: GO:0004713; protein tyrosine kinase activity
GO:0004992; platelet activating factor receptor activity
GO:0005017; platelet-derived growth factor-activated receptor activity
GO:0005019; platelet-derived growth factor beta-receptor activity
GO:0005088; Ras guanyl-nucleotide exchange factor activity
GO:0005102; receptor binding
GO:0005161; platelet-derived growth factor receptor binding
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0019899; enzyme binding
GO:0019901; protein kinase binding
GO:0038085; vascular endothelial growth factor binding
GO:0043548; phosphatidylinositol 3-kinase binding
GO:0046934; phosphatidylinositol-4,5-bisphosphate 3-kinase activity
GO:0048407; platelet-derived growth factor binding
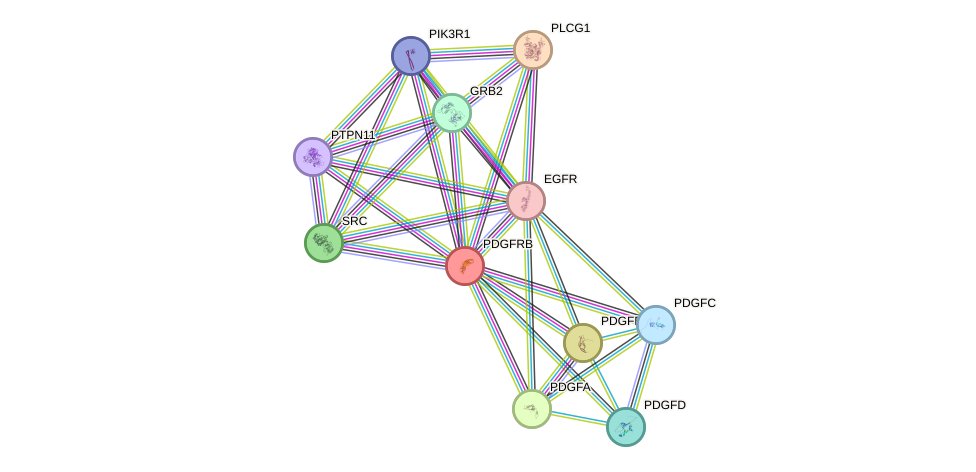
Protein interactions and network
- Protein-protein interacting partners in GenAge
- SHC1, PTPN11, STAT3, STAT5A, EGFR, PDGFB, PDGFRB, PTEN, HSP90AA1, GRB2, PIK3R1, PDGFRA
- STRING interaction network