GenAge entry for DDIT3 (Homo sapiens)
Gene name (HAGRID: 203)
- HGNC symbol
- DDIT3
- Aliases
- CHOP10; GADD153; CHOP
- Common name
- DNA-damage-inducible transcript 3
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on indirect or inconclusive evidence linking the gene product to ageing in humans or in one or more model systems
- Description
Also called GADD153 and CHOP, DDIT3 prevents transcription in response to DNA damage [880]. It also appears to be related to apoptosis. Although DDIT3 levels may increase with age [879], its relation to human ageing remains undetermined.
Cytogenetic information
- Cytogenetic band
- 12q13.1-q1
- Location
- 57,516,588 bp to 57,520,517 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0001955; blood vessel maturation
GO:0006355; regulation of transcription, DNA-templated
GO:0006974; cellular response to DNA damage stimulus
GO:0006983; ER overload response
GO:0006986; response to unfolded protein
GO:0007050; cell cycle arrest
GO:0016055; Wnt signaling pathway
GO:0032757; positive regulation of interleukin-8 production
GO:0032792; negative regulation of CREB transcription factor activity
GO:0034976; response to endoplasmic reticulum stress
GO:0036499; PERK-mediated unfolded protein response
GO:0036500; ATF6-mediated unfolded protein response
GO:0042594; response to starvation
GO:0042789; mRNA transcription from RNA polymerase II promoter
GO:0043161; proteasome-mediated ubiquitin-dependent protein catabolic process
GO:0043433; negative regulation of sequence-specific DNA binding transcription factor activity
GO:0043525; positive regulation of neuron apoptotic process
GO:0043620; regulation of DNA-templated transcription in response to stress
GO:0044324; regulation of transcription involved in anterior/posterior axis specification
GO:0045454; cell redox homeostasis
GO:0045599; negative regulation of fat cell differentiation
GO:0045662; negative regulation of myoblast differentiation
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0051209; release of sequestered calcium ion into cytosol
GO:0051898; negative regulation of protein kinase B signaling
GO:0070059; intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress
GO:0072655; establishment of protein localization to mitochondrion
GO:0090090; negative regulation of canonical Wnt signaling pathway
GO:1902237; positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway
GO:1903026; negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding
GO:1990440; positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress
GO:1990442; intrinsic apoptotic signaling pathway in response to nitrosative stress
GO:2000016; negative regulation of determination of dorsal identity
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005770; late endosome
GO:0005829; cytosol
GO:0032993; protein-DNA complex
GO:0035976; transcription factor AP-1 complex
GO:0036488; CHOP-C/EBP complex
GO:1990617; CHOP-ATF4 complex
GO:1990622; CHOP-ATF3 complex
Hide GO termsFunction: GO:0000976; transcription regulatory region sequence-specific DNA binding
GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0003677; DNA binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0003714; transcription corepressor activity
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0008140; cAMP response element binding protein binding
GO:0042803; protein homodimerization activity
GO:0043522; leucine zipper domain binding
GO:0044212; transcription regulatory region DNA binding
GO:0046982; protein heterodimerization activity
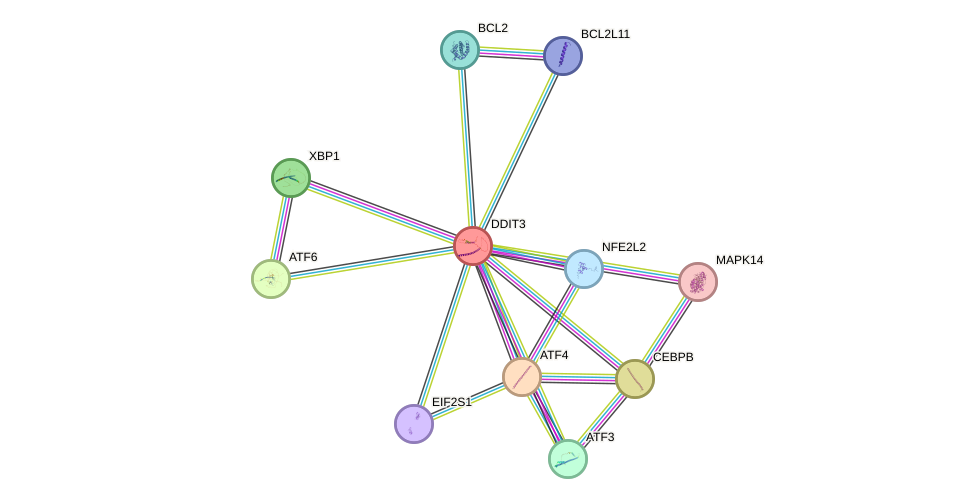
Protein interactions and network
- Protein-protein interacting partners in GenAge
- JUND, FOS, CREBBP, CEBPB, EP300, HDAC1, MAPK14, JUN, ATF2, DDIT3
- STRING interaction network