GenAge entry for HSPA1A (Homo sapiens)
Entry selected based on evidence directly linking the gene product to ageing in a mammalian model organism
Gene name (HAGRID: 160)
- HGNC symbol
- HSPA1A
- Aliases
- HSP70-1; HSPA1
- Common name
- heat shock 70kDa protein 1A
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence directly linking the gene product to ageing in a mammalian model organism
- Description
The HSPA1A protein, also known as hsp72 or hsp70, is a stress-induced chaperone [757]. Results from invertebrates have suggested that heat-shock proteins, such as HSPA1A, are involved in ageing. Overexpression of hsp70, a homologue of HSPA1A, extends lifespan in fruit flies [210]. Induction of HSPA1A after ischemia is diminished in aged rat hearts [908]. Overexpression of HSPA1A in mice with spinocerebellar ataxia type 1 offered protection against this disease, suggesting HSPA1A could protect against neurodegeneration [755]. However, in a more recent study, overexpression of hsp70 in mice was associated with growth retardation, tumor formation, and early death [1978]. Hsp70 basal levels show tissue-specific age-associated variations and are preserved in long-lived mice. Hsp70 levels from long-lived mice are similar to those found in adult WT mice. In adult PAM mice, which normally show accelerated aging, hsp70 levels are similar to those in chronologically old animals [4364]. Whether HSPA1A is related to human ageing remains to be determined.
Cytogenetic information
- Cytogenetic band
- 6p21.3
- Location
- 31,815,514 bp to 31,817,942 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0010628; positive regulation of gene expression
GO:0010941; regulation of cell death
GO:0030512; negative regulation of transforming growth factor beta receptor signaling pathway
GO:0031396; regulation of protein ubiquitination
GO:0031397; negative regulation of protein ubiquitination
GO:0032436; positive regulation of proteasomal ubiquitin-dependent protein catabolic process
GO:0032757; positive regulation of interleukin-8 production
GO:0034599; cellular response to oxidative stress
GO:0034605; cellular response to heat
GO:0042026; protein refolding
GO:0043488; regulation of mRNA stability
GO:0046034; ATP metabolic process
GO:0046718; viral entry into host cell
GO:0050821; protein stabilization
GO:0051092; positive regulation of NF-kappaB transcription factor activity
GO:0060548; negative regulation of cell death
GO:0070370; cellular heat acclimation
GO:0070434; positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway
GO:0090084; negative regulation of inclusion body assembly
GO:0098609; cell-cell adhesion
GO:1900034; regulation of cellular response to heat
GO:1901029; negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway
GO:1902236; negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway
GO:1902380; positive regulation of endoribonuclease activity
GO:1903265; positive regulation of tumor necrosis factor-mediated signaling pathway
GO:1904722; positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response
GO:2001240; negative regulation of extrinsic apoptotic signaling pathway in absence of ligand
Cellular component: GO:0000151; ubiquitin ligase complex
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005739; mitochondrion
GO:0005814; centriole
GO:0005829; cytosol
GO:0005913; cell-cell adherens junction
GO:0005925; focal adhesion
GO:0016234; inclusion body
GO:0048471; perinuclear region of cytoplasm
GO:0072562; blood microparticle
Hide GO termsFunction: GO:0001618; virus receptor activity
GO:0001664; G-protein coupled receptor binding
GO:0005102; receptor binding
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0016887; ATPase activity
GO:0019899; enzyme binding
GO:0031072; heat shock protein binding
GO:0031625; ubiquitin protein ligase binding
GO:0042623; ATPase activity, coupled
GO:0042826; histone deacetylase binding
GO:0044183; protein binding involved in protein folding
GO:0051082; unfolded protein binding
GO:0055131; C3HC4-type RING finger domain binding
GO:0098641; cadherin binding involved in cell-cell adhesion
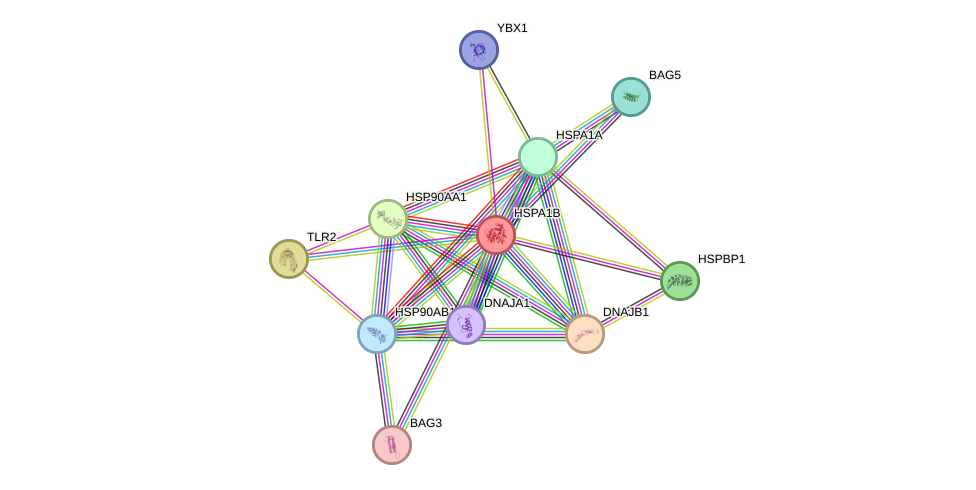
Protein interactions and network
- Protein-protein interacting partners in GenAge
- SHC1, TP53, TERT, MYC, EGFR, FOS, PTEN, BCL2, VCP, HSP90AA1, NR3C1, STK11, AR, EMD, HSF1, AIFM1, RELA, HSPD1, MAP3K5, BMI1, EEF1A1, HSPA8, MAPT, MAX, H2AFX, STUB1, CDKN1A, SPRTN
- STRING interaction network
Retrieve sequences for HSPA1A
Homologs in model organisms
No homologs found