GenAge entry for HTRA2 (Homo sapiens)
Entry selected based on evidence directly linking the gene product to ageing in a mammalian model organism
Gene name (HAGRID: 294)
- HGNC symbol
- HTRA2
- Aliases
- OMI; PARK13; PRSS25
- Common name
- HtrA serine peptidase 2
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence directly linking the gene product to ageing in a mammalian model organism
- Description
HTRA2 is a serine protease involved in apoptosis. Mice mutant for in Htra2 develop neurodegeneration 30–40 days after birth. However, also expressing human HTRA2 in the central nervous system rescues mice from the neurodegeneration and prevents premature death; the mice then go on to develop signs of premature ageing, including premature weight loss, hair loss, reduced fertility and kyphosis [3254]. These results suggest that HTRA2 might play a role in ageing beyond neurodegeneration, though further studies are necessary to establish this.
Cytogenetic information
- Cytogenetic band
- 2p12
- Location
- 74,529,405 bp to 74,533,556 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0006508; proteolysis
GO:0006672; ceramide metabolic process
GO:0007005; mitochondrion organization
GO:0007568; aging
GO:0007628; adult walking behavior
GO:0008630; intrinsic apoptotic signaling pathway in response to DNA damage
GO:0009635; response to herbicide
GO:0010822; positive regulation of mitochondrion organization
GO:0010942; positive regulation of cell death
GO:0016540; protein autoprocessing
GO:0019742; pentacyclic triterpenoid metabolic process
GO:0030900; forebrain development
GO:0034599; cellular response to oxidative stress
GO:0034605; cellular response to heat
GO:0035458; cellular response to interferon-beta
GO:0040014; regulation of multicellular organism growth
GO:0043065; positive regulation of apoptotic process
GO:0043280; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process
GO:0044257; cellular protein catabolic process
GO:0045786; negative regulation of cell cycle
GO:0048666; neuron development
GO:0071300; cellular response to retinoic acid
GO:0071363; cellular response to growth factor stimulus
GO:0097194; execution phase of apoptosis
GO:1901215; negative regulation of neuron death
GO:1902176; negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway
GO:1903146; regulation of mitophagy
GO:1903955; positive regulation of protein targeting to mitochondrion
GO:1904924; negative regulation of mitophagy in response to mitochondrial depolarization
GO:2001241; positive regulation of extrinsic apoptotic signaling pathway in absence of ligand
GO:2001269; positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway
Cellular component: GO:0000785; chromatin
GO:0005634; nucleus
GO:0005739; mitochondrion
GO:0005758; mitochondrial intermembrane space
GO:0005783; endoplasmic reticulum
GO:0005789; endoplasmic reticulum membrane
GO:0005829; cytosol
GO:0005856; cytoskeleton
GO:0009898; cytoplasmic side of plasma membrane
GO:0016020; membrane
GO:0031966; mitochondrial membrane
GO:0035631; CD40 receptor complex
Hide GO termsFunction: GO:0004252; serine-type endopeptidase activity
GO:0005515; protein binding
GO:0008233; peptidase activity
GO:0008236; serine-type peptidase activity
GO:0051082; unfolded protein binding
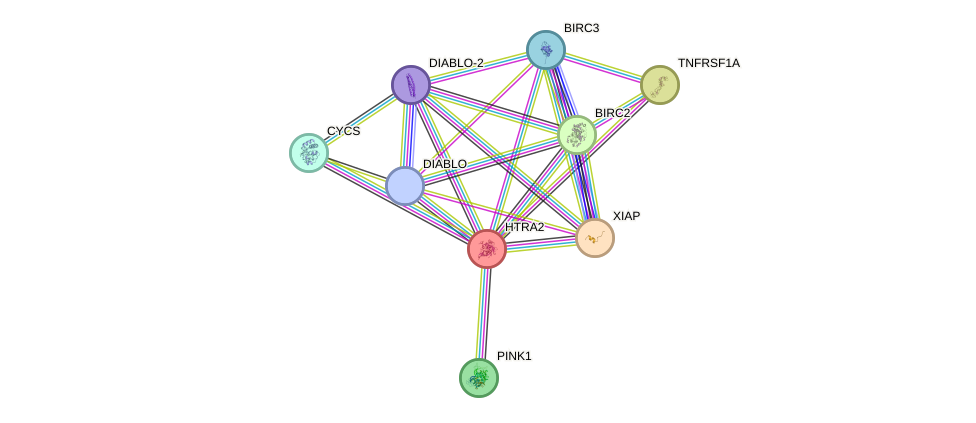
Protein interactions and network
- Protein-protein interacting partners in GenAge
- VCP, MAPK14, MAPK3, EEF1A1, CDK1, HTRA2
- STRING interaction network
Retrieve sequences for HTRA2
Homologs in model organisms
In other databases
- GenAge model organism genes
- A homolog of this gene for Mus musculus is present as Htra2