GenAge entry for PTEN (Homo sapiens)
Gene name (HAGRID: 63)
- HGNC symbol
- PTEN
- Aliases
- MMAC1; TEP1; PTEN1; BZS; MHAM
- Common name
- phosphatase and tensin homolog
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence directly linking the gene product to ageing in a non-mammalian animal model
- Description
PTEN is a tumour suppressor involved in cell cycle progression as an inhibitor of insulin (INS) signalling. In roundworms, a PTEN homologue has been related to development and longevity regulation through the INS-like pathway [1348]. The same gene has been shown to mediate nutrient-dependent cell cycle arrest and growth in the germline of roundworms [1771]. PTEN-null mice die at embryonic stages while heterozygous animals develop tumours [1757]. In mice lacking PTEN in oocytes the entire primordial follicle pool becomes activated and thus all primordial follicles become depleted in early adulthood causing premature ovarian failure [1905]. Mice carrying additional copies of PTEN are protected from cancer and present a modest (16%) extension of life span that is independent of their lower cancer incidence [2457]. Mutations in the human PTEN gene have been associated with cancer [1577], and further studies are necessary to determine whether PTEN plays a role in human ageing.
Cytogenetic information
- Cytogenetic band
- 10q23.3
- Location
- 87,863,438 bp to 87,971,930 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000079; regulation of cyclin-dependent protein serine/threonine kinase activity
GO:0001525; angiogenesis
GO:0001933; negative regulation of protein phosphorylation
GO:0002902; regulation of B cell apoptotic process
GO:0006470; protein dephosphorylation
GO:0006661; phosphatidylinositol biosynthetic process
GO:0006915; apoptotic process
GO:0007270; neuron-neuron synaptic transmission
GO:0007416; synapse assembly
GO:0007417; central nervous system development
GO:0007507; heart development
GO:0007568; aging
GO:0007584; response to nutrient
GO:0007611; learning or memory
GO:0007613; memory
GO:0007626; locomotory behavior
GO:0008283; cell proliferation
GO:0008284; positive regulation of cell proliferation
GO:0008285; negative regulation of cell proliferation
GO:0009749; response to glucose
GO:0010043; response to zinc ion
GO:0010975; regulation of neuron projection development
GO:0014067; negative regulation of phosphatidylinositol 3-kinase signaling
GO:0016477; cell migration
GO:0021542; dentate gyrus development
GO:0021955; central nervous system neuron axonogenesis
GO:0030336; negative regulation of cell migration
GO:0030534; adult behavior
GO:0031642; negative regulation of myelination
GO:0031647; regulation of protein stability
GO:0031658; negative regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle
GO:0032228; regulation of synaptic transmission, GABAergic
GO:0032286; central nervous system myelin maintenance
GO:0032355; response to estradiol
GO:0032535; regulation of cellular component size
GO:0033032; regulation of myeloid cell apoptotic process
GO:0033198; response to ATP
GO:0033555; multicellular organismal response to stress
GO:0035176; social behavior
GO:0035335; peptidyl-tyrosine dephosphorylation
GO:0042493; response to drug
GO:0042711; maternal behavior
GO:0043066; negative regulation of apoptotic process
GO:0043491; protein kinase B signaling
GO:0043542; endothelial cell migration
GO:0043647; inositol phosphate metabolic process
GO:0045471; response to ethanol
GO:0045475; locomotor rhythm
GO:0045792; negative regulation of cell size
GO:0046621; negative regulation of organ growth
GO:0046685; response to arsenic-containing substance
GO:0046855; inositol phosphate dephosphorylation
GO:0046856; phosphatidylinositol dephosphorylation
GO:0048008; platelet-derived growth factor receptor signaling pathway
GO:0048015; phosphatidylinositol-mediated signaling
GO:0048681; negative regulation of axon regeneration
GO:0048738; cardiac muscle tissue development
GO:0048853; forebrain morphogenesis
GO:0048854; brain morphogenesis
GO:0050680; negative regulation of epithelial cell proliferation
GO:0050765; negative regulation of phagocytosis
GO:0050771; negative regulation of axonogenesis
GO:0050821; protein stabilization
GO:0050852; T cell receptor signaling pathway
GO:0051091; positive regulation of sequence-specific DNA binding transcription factor activity
GO:0051895; negative regulation of focal adhesion assembly
GO:0051898; negative regulation of protein kinase B signaling
GO:0060024; rhythmic synaptic transmission
GO:0060044; negative regulation of cardiac muscle cell proliferation
GO:0060070; canonical Wnt signaling pathway
GO:0060074; synapse maturation
GO:0060134; prepulse inhibition
GO:0060179; male mating behavior
GO:0060291; long-term synaptic potentiation
GO:0060292; long term synaptic depression
GO:0060736; prostate gland growth
GO:0060997; dendritic spine morphogenesis
GO:0061002; negative regulation of dendritic spine morphogenesis
GO:0070373; negative regulation of ERK1 and ERK2 cascade
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:0071456; cellular response to hypoxia
GO:0090071; negative regulation of ribosome biogenesis
GO:0090344; negative regulation of cell aging
GO:0090394; negative regulation of excitatory postsynaptic potential
GO:0097105; presynaptic membrane assembly
GO:0097107; postsynaptic density assembly
GO:1903984; positive regulation of TRAIL-activated apoptotic signaling pathway
GO:1904668; positive regulation of ubiquitin protein ligase activity
GO:2000060; positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process
GO:2000134; negative regulation of G1/S transition of mitotic cell cycle
GO:2000463; positive regulation of excitatory postsynaptic potential
GO:2000808; negative regulation of synaptic vesicle clustering
Cellular component: GO:0005576; extracellular region
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005739; mitochondrion
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0009898; cytoplasmic side of plasma membrane
GO:0016324; apical plasma membrane
GO:0016605; PML body
GO:0035749; myelin sheath adaxonal region
GO:0042995; cell projection
GO:0043005; neuron projection
GO:0043197; dendritic spine
GO:0043220; Schmidt-Lanterman incisure
GO:0045211; postsynaptic membrane
Hide GO termsFunction: GO:0000287; magnesium ion binding
GO:0004438; phosphatidylinositol-3-phosphatase activity
GO:0004721; phosphoprotein phosphatase activity
GO:0004722; protein serine/threonine phosphatase activity
GO:0004725; protein tyrosine phosphatase activity
GO:0005161; platelet-derived growth factor receptor binding
GO:0005515; protein binding
GO:0008138; protein tyrosine/serine/threonine phosphatase activity
GO:0008289; lipid binding
GO:0010997; anaphase-promoting complex binding
GO:0016314; phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity
GO:0019899; enzyme binding
GO:0019901; protein kinase binding
GO:0030165; PDZ domain binding
GO:0042802; identical protein binding
GO:0051717; inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity
GO:0051800; phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity
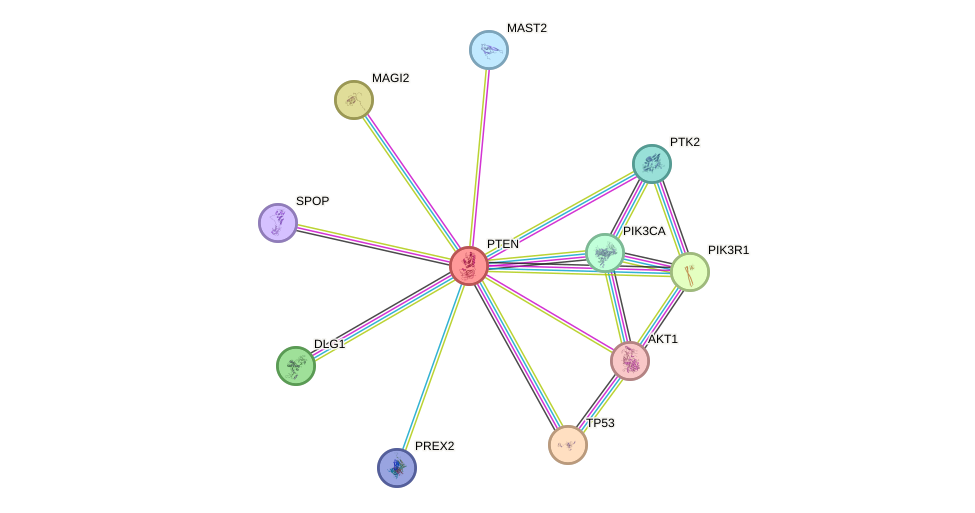
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, TXN, AKT1, PDGFRB, HSP90AA1, EGR1, UBE2I, EEF2, AR, PRDX1, HSPD1, HSPA1A, PTK2B, PTK2, BMI1, HSPA8, MXD1, ESR1, CTNNB1, PPP1CA, DBN1, STUB1, PDGFRA
- STRING interaction network
Retrieve sequences for PTEN
Homologs in model organisms
In other databases
- GenAge model organism genes
- CellAge
- This gene is present as PTEN