GenAge entry for PTK2B (Homo sapiens)
Gene name (HAGRID: 165)
- HGNC symbol
- PTK2B
- Aliases
- CAKB; PYK2; RAFTK; PTK; CADTK; FAK2
- Common name
- protein tyrosine kinase 2 beta
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
Involved in stress response and signal transduction, PTK2B is an important player in a variety of processes including the regulation of ion channels by calcium and MAPK signalling [769]. PTK2B is a focal adhesion kinase (FAK), which could be involved in cellular senescence and apoptosis [1716]. Its role, if any, in human ageing is unknown, but PTK2B could potentially be involved in downstream signalling cascades related to ageing.
Cytogenetic information
- Cytogenetic band
- 8p21.1
- Location
- 27,311,482 bp to 27,459,391 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000165; MAPK cascade
GO:0001525; angiogenesis
GO:0001556; oocyte maturation
GO:0001666; response to hypoxia
GO:0001954; positive regulation of cell-matrix adhesion
GO:0002040; sprouting angiogenesis
GO:0002250; adaptive immune response
GO:0002315; marginal zone B cell differentiation
GO:0006461; protein complex assembly
GO:0006468; protein phosphorylation
GO:0006915; apoptotic process
GO:0006950; response to stress
GO:0006968; cellular defense response
GO:0006970; response to osmotic stress
GO:0007165; signal transduction
GO:0007166; cell surface receptor signaling pathway
GO:0007172; signal complex assembly
GO:0007173; epidermal growth factor receptor signaling pathway
GO:0007204; positive regulation of cytosolic calcium ion concentration
GO:0007229; integrin-mediated signaling pathway
GO:0008284; positive regulation of cell proliferation
GO:0008285; negative regulation of cell proliferation
GO:0008360; regulation of cell shape
GO:0009612; response to mechanical stimulus
GO:0009725; response to hormone
GO:0009749; response to glucose
GO:0010226; response to lithium ion
GO:0010595; positive regulation of endothelial cell migration
GO:0010656; negative regulation of muscle cell apoptotic process
GO:0010752; regulation of cGMP-mediated signaling
GO:0010758; regulation of macrophage chemotaxis
GO:0010976; positive regulation of neuron projection development
GO:0014009; glial cell proliferation
GO:0018108; peptidyl-tyrosine phosphorylation
GO:0030155; regulation of cell adhesion
GO:0030307; positive regulation of cell growth
GO:0030335; positive regulation of cell migration
GO:0030502; negative regulation of bone mineralization
GO:0030826; regulation of cGMP biosynthetic process
GO:0030838; positive regulation of actin filament polymerization
GO:0031175; neuron projection development
GO:0032960; regulation of inositol trisphosphate biosynthetic process
GO:0033209; tumor necrosis factor-mediated signaling pathway
GO:0035235; ionotropic glutamate receptor signaling pathway
GO:0035902; response to immobilization stress
GO:0038083; peptidyl-tyrosine autophosphorylation
GO:0042220; response to cocaine
GO:0042493; response to drug
GO:0042542; response to hydrogen peroxide
GO:0042976; activation of Janus kinase activity
GO:0043066; negative regulation of apoptotic process
GO:0043149; stress fiber assembly
GO:0043267; negative regulation of potassium ion transport
GO:0043507; positive regulation of JUN kinase activity
GO:0043524; negative regulation of neuron apoptotic process
GO:0043534; blood vessel endothelial cell migration
GO:0043552; positive regulation of phosphatidylinositol 3-kinase activity
GO:0045087; innate immune response
GO:0045429; positive regulation of nitric oxide biosynthetic process
GO:0045453; bone resorption
GO:0045471; response to ethanol
GO:0045638; negative regulation of myeloid cell differentiation
GO:0045727; positive regulation of translation
GO:0045766; positive regulation of angiogenesis
GO:0045860; positive regulation of protein kinase activity
GO:0046330; positive regulation of JNK cascade
GO:0046777; protein autophosphorylation
GO:0048010; vascular endothelial growth factor receptor signaling pathway
GO:0048041; focal adhesion assembly
GO:0050731; positive regulation of peptidyl-tyrosine phosphorylation
GO:0050848; regulation of calcium-mediated signaling
GO:0051000; positive regulation of nitric-oxide synthase activity
GO:0051279; regulation of release of sequestered calcium ion into cytosol
GO:0051591; response to cAMP
GO:0051592; response to calcium ion
GO:0051968; positive regulation of synaptic transmission, glutamatergic
GO:0060291; long-term synaptic potentiation
GO:0060292; long term synaptic depression
GO:0070098; chemokine-mediated signaling pathway
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:0071300; cellular response to retinoic acid
GO:0071498; cellular response to fluid shear stress
GO:0090630; activation of GTPase activity
GO:2000058; regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process
GO:2000060; positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process
GO:2000114; regulation of establishment of cell polarity
GO:2000249; regulation of actin cytoskeleton reorganization
GO:2000310; regulation of N-methyl-D-aspartate selective glutamate receptor activity
GO:2000463; positive regulation of excitatory postsynaptic potential
GO:2000538; positive regulation of B cell chemotaxis
GO:2000573; positive regulation of DNA biosynthetic process
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005829; cytosol
GO:0005856; cytoskeleton
GO:0005925; focal adhesion
GO:0005938; cell cortex
GO:0014069; postsynaptic density
GO:0017146; NMDA selective glutamate receptor complex
GO:0030027; lamellipodium
GO:0030424; axon
GO:0030425; dendrite
GO:0030426; growth cone
GO:0031234; extrinsic component of cytoplasmic side of plasma membrane
GO:0043025; neuronal cell body
GO:0044297; cell body
GO:0045121; membrane raft
GO:0048471; perinuclear region of cytoplasm
GO:0097440; apical dendrite
Hide GO termsFunction: GO:0004683; calmodulin-dependent protein kinase activity
GO:0004713; protein tyrosine kinase activity
GO:0004715; non-membrane spanning protein tyrosine kinase activity
GO:0004871; signal transducer activity
GO:0004972; NMDA glutamate receptor activity
GO:0005102; receptor binding
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0032403; protein complex binding
GO:0043423; 3-phosphoinositide-dependent protein kinase binding
Protein interactions and network
- Protein-protein interacting partners in GenAge
- PTPN11, IL7R, EGFR, ERBB2, PRKCD, PTEN, HSP90AA1, GRB2, PTK2B, MAPT, JAK2, SOCS2
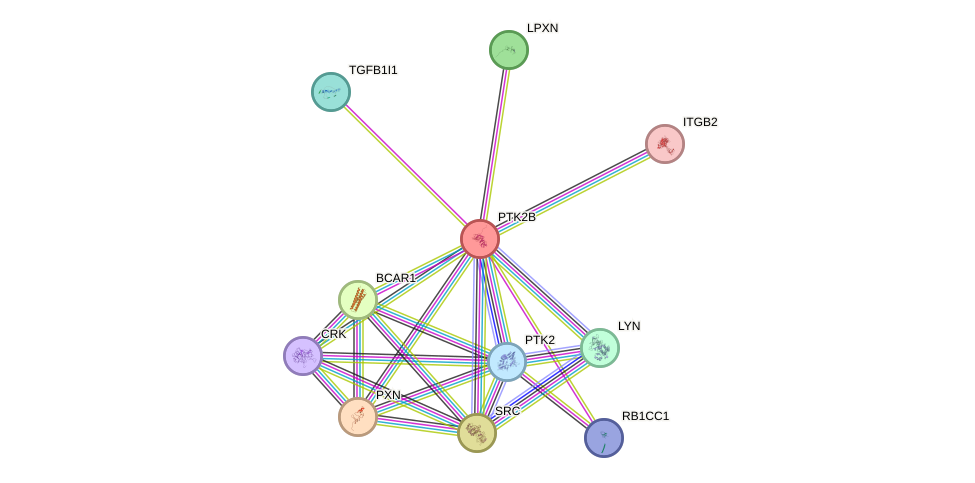
- STRING interaction network
Retrieve sequences for PTK2B
Homologs in model organisms
- Caenorhabditis elegans
- kin-32
- Danio rerio
- ptk2bb
- Drosophila melanogaster
- Fak
- Mus musculus
- Ptk2b
- Rattus norvegicus
- Ptk2b