GenAge entry for STAT5B (Homo sapiens)
Gene name (HAGRID: 21)
- HGNC symbol
- STAT5B
- Aliases
- Common name
- signal transducer and activator of transcription 5B
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
STAT5B is an important player in the activation of transcription and intracellular signalling. Mice with disrupted STAT5B and STAT5A, though short-lived due to numerous other deficiencies, exhibit phenotypes similar to those observed in GHR knockout mice in that they are 20%-40% smaller than controls with signs of disrupted GH1 functions such as lower IGF1 levels [1892]. Absence of Stat5 from pancreatic beta-cells in mice has no discernable impact on islet development and function in young animals, however, Stat5 appears to be an important component in beta-cell function in older mice [3666]. Its relation to other proteins that may be involved in ageing make STAT5B a possible, albeit unproven, player in human ageing and age-related disease [266].
Cytogenetic information
- Cytogenetic band
- 17q11.2
- Location
- 42,199,177 bp to 42,276,406 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000255; allantoin metabolic process
GO:0001553; luteinization
GO:0001779; natural killer cell differentiation
GO:0006101; citrate metabolic process
GO:0006103; 2-oxoglutarate metabolic process
GO:0006105; succinate metabolic process
GO:0006107; oxaloacetate metabolic process
GO:0006357; regulation of transcription from RNA polymerase II promoter
GO:0006366; transcription from RNA polymerase II promoter
GO:0006549; isoleucine metabolic process
GO:0006573; valine metabolic process
GO:0006600; creatine metabolic process
GO:0006631; fatty acid metabolic process
GO:0007259; JAK-STAT cascade
GO:0007565; female pregnancy
GO:0007595; lactation
GO:0018108; peptidyl-tyrosine phosphorylation
GO:0019218; regulation of steroid metabolic process
GO:0019530; taurine metabolic process
GO:0019915; lipid storage
GO:0030856; regulation of epithelial cell differentiation
GO:0032355; response to estradiol
GO:0032819; positive regulation of natural killer cell proliferation
GO:0032825; positive regulation of natural killer cell differentiation
GO:0032870; cellular response to hormone stimulus
GO:0033077; T cell differentiation in thymus
GO:0038161; prolactin signaling pathway
GO:0040014; regulation of multicellular organism growth
GO:0040018; positive regulation of multicellular organism growth
GO:0042104; positive regulation of activated T cell proliferation
GO:0042448; progesterone metabolic process
GO:0043029; T cell homeostasis
GO:0043066; negative regulation of apoptotic process
GO:0045086; positive regulation of interleukin-2 biosynthetic process
GO:0045579; positive regulation of B cell differentiation
GO:0045588; positive regulation of gamma-delta T cell differentiation
GO:0045647; negative regulation of erythrocyte differentiation
GO:0045648; positive regulation of erythrocyte differentiation
GO:0045931; positive regulation of mitotic cell cycle
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0045954; positive regulation of natural killer cell mediated cytotoxicity
GO:0046449; creatinine metabolic process
GO:0046543; development of secondary female sexual characteristics
GO:0046544; development of secondary male sexual characteristics
GO:0048541; Peyer's patch development
GO:0050729; positive regulation of inflammatory response
GO:0060397; JAK-STAT cascade involved in growth hormone signaling pathway
GO:0070669; response to interleukin-2
GO:0070670; response to interleukin-4
GO:0070672; response to interleukin-15
GO:0071363; cellular response to growth factor stimulus
GO:0071364; cellular response to epidermal growth factor stimulus
GO:0097531; mast cell migration
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005829; cytosol
Hide GO termsFunction: GO:0000979; RNA polymerase II core promoter sequence-specific DNA binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0003682; chromatin binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0004713; protein tyrosine kinase activity
GO:0004871; signal transducer activity
GO:0005515; protein binding
GO:0019903; protein phosphatase binding
GO:0035259; glucocorticoid receptor binding
GO:0046983; protein dimerization activity
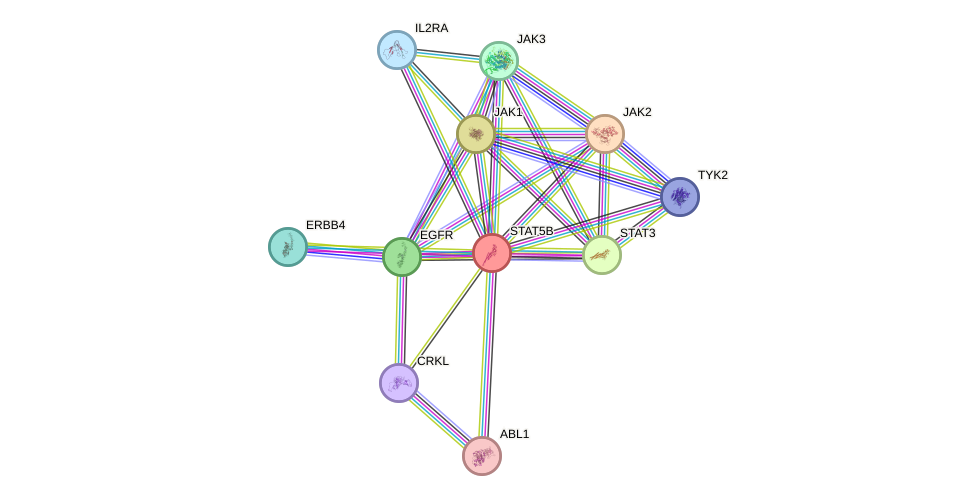
Protein interactions and network
- Protein-protein interacting partners in GenAge
- GHR, SHC1, PTPN11, STAT5B, STAT3, PTPN1, EGFR, INSR, NR3C1, APP, JAK2
- STRING interaction network
Retrieve sequences for STAT5B
Homologs in model organisms
In other databases
- CellAge
- This gene is present as STAT5B