GenAge entry for APOE (Homo sapiens)
Gene name (HAGRID: 138)
- HGNC symbol
- APOE
- Aliases
- AD2
- Common name
- apolipoprotein E
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene or its product to human longevity and/or multiple age-related phenotypes
- Description
APOE is involved in lipid metabolism. Mice without APOE show dysregulations in lipid metabolism, having higher plasma cholesterol levels and developing arterial lesions [699], displaying DNA oxidative stress at a hepatic level [3602], and presenting severe atherosclerosis and cutaneous xanthomatosis [2138]. They also have a significantly shorter lifespan than wild-type [2138]. Neurodegeneration has also been reported in APOE-null mice [1699].
Several polymorphisms in the human APOE gene have been associated with diseases, such as increased risk of myocardial infarction [1700] and Alzheimer’s disease [4497]. Polymorphisms in APOE have been linked to human longevity [145]. There are four isoforms of APOE in humans. In Alzheimer’s disease the ApoE2 isoform is protective and the ApoE4 isoform is causative and stimulates amyloid beta production [4497]. ApoE4 also exacerbates tauopathy in mouse models of Alzheimer’s disease and ApoE4 knock-in mice show significantly higher levels of tau in the brain [4498]. Despite its association with age-related disease the ApoE4 isoform is the second most common after the ApoE3 isoform and is the ancestral form of APOE [4499]. Clearly, APOE has an impact on various age-related diseases, such as atherosclerosis and neurodegeneration, but its overall impact on the human ageing process remains to be determined.
Cytogenetic information
- Cytogenetic band
- 19q13.2
- Location
- 44,905,748 bp to 44,909,395 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000302; response to reactive oxygen species
GO:0001523; retinoid metabolic process
GO:0001937; negative regulation of endothelial cell proliferation
GO:0002021; response to dietary excess
GO:0006641; triglyceride metabolic process
GO:0006707; cholesterol catabolic process
GO:0006874; cellular calcium ion homeostasis
GO:0006898; receptor-mediated endocytosis
GO:0007010; cytoskeleton organization
GO:0007186; G-protein coupled receptor signaling pathway
GO:0007263; nitric oxide mediated signal transduction
GO:0007271; synaptic transmission, cholinergic
GO:0008203; cholesterol metabolic process
GO:0010468; regulation of gene expression
GO:0010544; negative regulation of platelet activation
GO:0010873; positive regulation of cholesterol esterification
GO:0010875; positive regulation of cholesterol efflux
GO:0010877; lipid transport involved in lipid storage
GO:0015909; long-chain fatty acid transport
GO:0017038; protein import
GO:0019068; virion assembly
GO:0019433; triglyceride catabolic process
GO:0019934; cGMP-mediated signaling
GO:0030195; negative regulation of blood coagulation
GO:0030516; regulation of axon extension
GO:0030828; positive regulation of cGMP biosynthetic process
GO:0031102; neuron projection regeneration
GO:0032489; regulation of Cdc42 protein signal transduction
GO:0032805; positive regulation of low-density lipoprotein particle receptor catabolic process
GO:0033344; cholesterol efflux
GO:0033700; phospholipid efflux
GO:0034372; very-low-density lipoprotein particle remodeling
GO:0034374; low-density lipoprotein particle remodeling
GO:0034375; high-density lipoprotein particle remodeling
GO:0034380; high-density lipoprotein particle assembly
GO:0034382; chylomicron remnant clearance
GO:0034384; high-density lipoprotein particle clearance
GO:0034447; very-low-density lipoprotein particle clearance
GO:0042157; lipoprotein metabolic process
GO:0042158; lipoprotein biosynthetic process
GO:0042159; lipoprotein catabolic process
GO:0042311; vasodilation
GO:0042632; cholesterol homeostasis
GO:0043407; negative regulation of MAP kinase activity
GO:0043524; negative regulation of neuron apoptotic process
GO:0043537; negative regulation of blood vessel endothelial cell migration
GO:0043691; reverse cholesterol transport
GO:0044794; positive regulation by host of viral process
GO:0045541; negative regulation of cholesterol biosynthetic process
GO:0046889; positive regulation of lipid biosynthetic process
GO:0046907; intracellular transport
GO:0048168; regulation of neuronal synaptic plasticity
GO:0048844; artery morphogenesis
GO:0050728; negative regulation of inflammatory response
GO:0051000; positive regulation of nitric-oxide synthase activity
GO:0051044; positive regulation of membrane protein ectodomain proteolysis
GO:0051651; maintenance of location in cell
GO:0055089; fatty acid homeostasis
GO:0060999; positive regulation of dendritic spine development
GO:0070328; triglyceride homeostasis
GO:0090090; negative regulation of canonical Wnt signaling pathway
GO:0090209; negative regulation of triglyceride metabolic process
GO:0097113; AMPA glutamate receptor clustering
GO:0097114; NMDA glutamate receptor clustering
GO:0098869; cellular oxidant detoxification
GO:1900221; regulation of beta-amyloid clearance
GO:1901215; negative regulation of neuron death
GO:1901628; positive regulation of postsynaptic membrane organization
GO:1901630; negative regulation of presynaptic membrane organization
GO:1902430; negative regulation of beta-amyloid formation
GO:1902952; positive regulation of dendritic spine maintenance
GO:1902995; positive regulation of phospholipid efflux
GO:1903002; positive regulation of lipid transport across blood brain barrier
Cellular component: GO:0005576; extracellular region
GO:0005615; extracellular space
GO:0005634; nucleus
GO:0005737; cytoplasm
GO:0005769; early endosome
GO:0005783; endoplasmic reticulum
GO:0005794; Golgi apparatus
GO:0005886; plasma membrane
GO:0016020; membrane
GO:0030425; dendrite
GO:0031012; extracellular matrix
GO:0034361; very-low-density lipoprotein particle
GO:0034362; low-density lipoprotein particle
GO:0034363; intermediate-density lipoprotein particle
GO:0034364; high-density lipoprotein particle
GO:0042627; chylomicron
GO:0043025; neuronal cell body
GO:0070062; extracellular exosome
GO:0071682; endocytic vesicle lumen
GO:0072562; blood microparticle
GO:1903561; extracellular vesicle
Hide GO termsFunction: GO:0001540; beta-amyloid binding
GO:0005319; lipid transporter activity
GO:0005515; protein binding
GO:0005543; phospholipid binding
GO:0008201; heparin binding
GO:0008289; lipid binding
GO:0015485; cholesterol binding
GO:0016209; antioxidant activity
GO:0017127; cholesterol transporter activity
GO:0042802; identical protein binding
GO:0042803; protein homodimerization activity
GO:0046911; metal chelating activity
GO:0048156; tau protein binding
GO:0050750; low-density lipoprotein particle receptor binding
GO:0060228; phosphatidylcholine-sterol O-acyltransferase activator activity
GO:0070326; very-low-density lipoprotein particle receptor binding
GO:0071813; lipoprotein particle binding
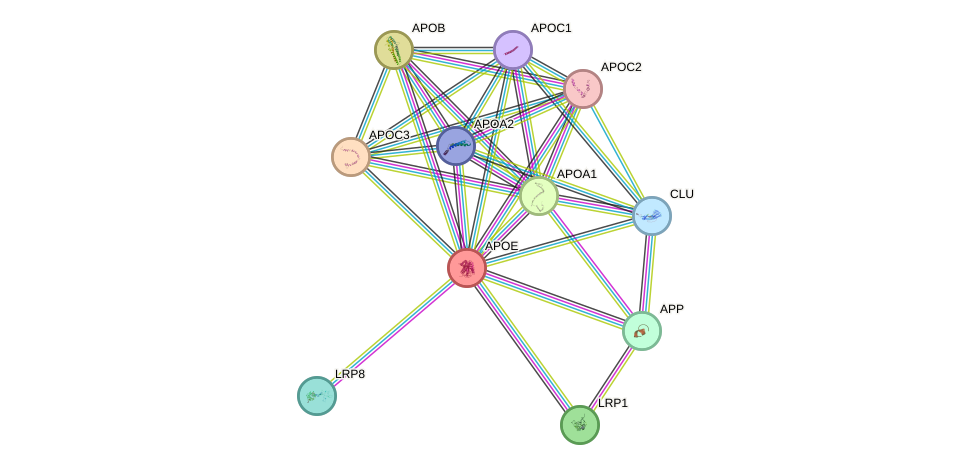
Protein interactions and network
- Protein-protein interacting partners in GenAge
- LRP2, APOE, A2M, PCMT1, MAPT, PSEN1
- STRING interaction network
Retrieve sequences for APOE
Homologs in model organisms
In other databases
- GenAge model organism genes
- A homolog of this gene for Mus musculus is present as Apoe
- LongevityMap
- This gene is present as APOE