GenAge entry for FAS (Homo sapiens)
Gene name (HAGRID: 115)
- HGNC symbol
- FAS
- Aliases
- CD95; APO-1; FAS1; APT1; TNFRSF6
- Common name
- Fas cell surface death receptor
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on indirect or inconclusive evidence linking the gene product to ageing in humans or in one or more model systems
- Description
FAS encodes multiple transcripts involved in apoptosis and development. It is also related to cancer progression. The expression level of FAS changes during ageing [937], and there are some results correlating FAS with age-related changes [1170]. Nonetheless, polymorphisms in human FAS have not been associated with human longevity [935].
Cytogenetic information
- Cytogenetic band
- 10q24.1
- Location
- 88,990,531 bp to 89,015,785 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0002377; immunoglobulin production
GO:0003014; renal system process
GO:0006461; protein complex assembly
GO:0006915; apoptotic process
GO:0006919; activation of cysteine-type endopeptidase activity involved in apoptotic process
GO:0006924; activation-induced cell death of T cells
GO:0006925; inflammatory cell apoptotic process
GO:0006954; inflammatory response
GO:0006955; immune response
GO:0007165; signal transduction
GO:0007420; brain development
GO:0007623; circadian rhythm
GO:0008625; extrinsic apoptotic signaling pathway via death domain receptors
GO:0009636; response to toxic substance
GO:0010467; gene expression
GO:0019724; B cell mediated immunity
GO:0032464; positive regulation of protein homooligomerization
GO:0032496; response to lipopolysaccharide
GO:0033209; tumor necrosis factor-mediated signaling pathway
GO:0042127; regulation of cell proliferation
GO:0042981; regulation of apoptotic process
GO:0043065; positive regulation of apoptotic process
GO:0043066; negative regulation of apoptotic process
GO:0043281; regulation of cysteine-type endopeptidase activity involved in apoptotic process
GO:0043410; positive regulation of MAPK cascade
GO:0045060; negative thymic T cell selection
GO:0045619; regulation of lymphocyte differentiation
GO:0045637; regulation of myeloid cell differentiation
GO:0048536; spleen development
GO:0050869; negative regulation of B cell activation
GO:0051260; protein homooligomerization
GO:0051384; response to glucocorticoid
GO:0070230; positive regulation of lymphocyte apoptotic process
GO:0071260; cellular response to mechanical stimulus
GO:0071285; cellular response to lithium ion
GO:0071455; cellular response to hyperoxia
GO:0097049; motor neuron apoptotic process
GO:0097190; apoptotic signaling pathway
GO:0097191; extrinsic apoptotic signaling pathway
GO:0097192; extrinsic apoptotic signaling pathway in absence of ligand
GO:0097284; hepatocyte apoptotic process
GO:0097296; activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway
GO:0097527; necroptotic signaling pathway
GO:1902041; regulation of extrinsic apoptotic signaling pathway via death domain receptors
GO:1902042; negative regulation of extrinsic apoptotic signaling pathway via death domain receptors
GO:2001241; positive regulation of extrinsic apoptotic signaling pathway in absence of ligand
Cellular component: GO:0005634; nucleus
GO:0005737; cytoplasm
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0005887; integral component of plasma membrane
GO:0009897; external side of plasma membrane
GO:0009986; cell surface
GO:0031264; death-inducing signaling complex
GO:0031265; CD95 death-inducing signaling complex
GO:0043005; neuron projection
GO:0045121; membrane raft
GO:0070062; extracellular exosome
Hide GO termsFunction: GO:0004871; signal transducer activity
GO:0004872; receptor activity
GO:0005031; tumor necrosis factor-activated receptor activity
GO:0005515; protein binding
GO:0019900; kinase binding
GO:0042802; identical protein binding
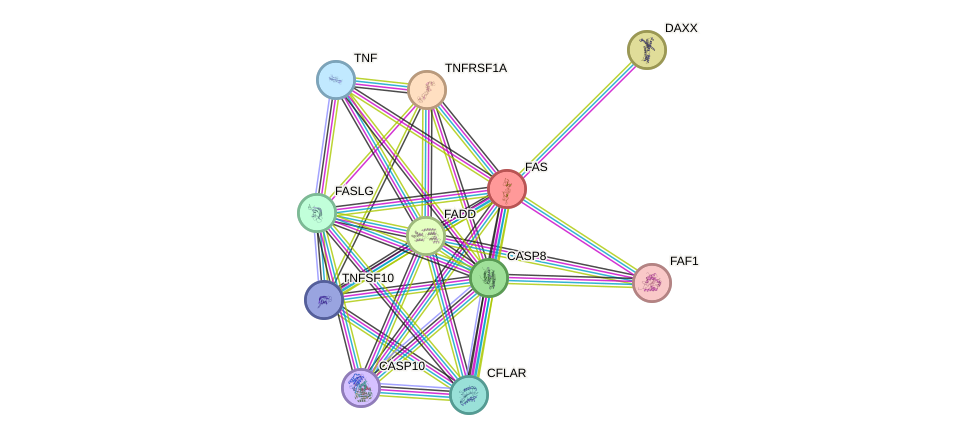
Protein interactions and network
- Protein-protein interacting partners in GenAge
- UBE2I, TNF, PML, PRKCA, FAS, RB1, MAPK8, MAP3K5, EEF1A1, SUMO1, CTNNB1, TP63, SQSTM1
- STRING interaction network