GenAge entry for CEBPA (Homo sapiens)
Gene name (HAGRID: 88)
- HGNC symbol
- CEBPA
- Aliases
- C/EBP-alpha; CEBP
- Common name
- CCAAT/enhancer binding protein (C/EBP), alpha
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
CEBPA is a transcription factor expressed in adipose tissues that modulates the expression of leptin (LEP). It can also affect the cell cycle. CEBPA-null mice die shortly after birth [1128]. Replacing the CEBPA gene by CEBPB in mice increases lifespan by about 20% [1122].
Cytogenetic information
- Cytogenetic band
- 19q13.1
- Location
- 33,299,934 bp to 33,302,564 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000050; urea cycle
GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0001889; liver development
GO:0001892; embryonic placenta development
GO:0006091; generation of precursor metabolites and energy
GO:0006351; transcription, DNA-templated
GO:0006366; transcription from RNA polymerase II promoter
GO:0007005; mitochondrion organization
GO:0007219; Notch signaling pathway
GO:0008203; cholesterol metabolic process
GO:0008285; negative regulation of cell proliferation
GO:0016032; viral process
GO:0019221; cytokine-mediated signaling pathway
GO:0030099; myeloid cell differentiation
GO:0030225; macrophage differentiation
GO:0030324; lung development
GO:0030851; granulocyte differentiation
GO:0032436; positive regulation of proteasomal ubiquitin-dependent protein catabolic process
GO:0042593; glucose homeostasis
GO:0045444; fat cell differentiation
GO:0045600; positive regulation of fat cell differentiation
GO:0045669; positive regulation of osteoblast differentiation
GO:0045736; negative regulation of cyclin-dependent protein serine/threonine kinase activity
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0045945; positive regulation of transcription from RNA polymerase III promoter
GO:0048469; cell maturation
GO:0048839; inner ear development
GO:0050872; white fat cell differentiation
GO:0050873; brown fat cell differentiation
GO:0055088; lipid homeostasis
GO:0071285; cellular response to lithium ion
GO:0071356; cellular response to tumor necrosis factor
GO:0071407; cellular response to organic cyclic compound
Cellular component: GO:0005634; nucleus
GO:0005730; nucleolus
GO:0090575; RNA polymerase II transcription factor complex
Hide GO termsFunction: GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001228; transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding
GO:0003677; DNA binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0003705; transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding
GO:0003713; transcription coactivator activity
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0019900; kinase binding
GO:0042803; protein homodimerization activity
GO:0044212; transcription regulatory region DNA binding
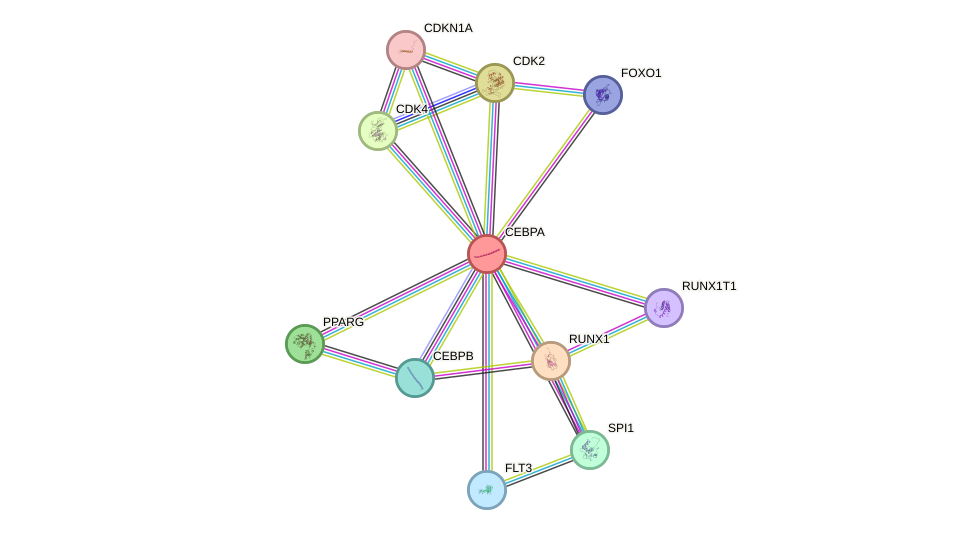
Protein interactions and network
- Protein-protein interacting partners in GenAge
- MYC, FOS, PARP1, NR3C1, CEBPA, CEBPB, AR, XRCC5, XRCC6, RB1, HDAC1, MAPK8, JUN, ATF2, HDAC2, MAX, ESR1, CDKN1A
- STRING interaction network
Retrieve sequences for CEBPA
Homologs in model organisms
In other databases
- GenAge model organism genes
- A homolog of this gene for Mus musculus is present as Cebpa