GenAge entry for PPARA (Homo sapiens)
Gene name (HAGRID: 55)
- HGNC symbol
- PPARA
- Aliases
- hPPAR; NR1C1; PPAR
- Common name
- peroxisome proliferator-activated receptor alpha
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on indirect or inconclusive evidence linking the gene product to ageing in humans or in one or more model systems
- Description
PPARA is a transcription factor that may be involved in the metabolic control of the expression of genes encoding fatty acid oxidation enzymes. Knockout mice have been reported to be protected from insulin (INS) resistance induced by high-fat diet [1928]. Certain age-related pathologies, such as heart disease, may be related to PPARA [392], but it unclear whether PPARA is a major player in human ageing.
Cytogenetic information
- Cytogenetic band
- 22q13.31
- Location
- 46,150,596 bp to 46,243,756 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0001666; response to hypoxia
GO:0006367; transcription initiation from RNA polymerase II promoter
GO:0006629; lipid metabolic process
GO:0006631; fatty acid metabolic process
GO:0007507; heart development
GO:0008544; epidermis development
GO:0010745; negative regulation of macrophage derived foam cell differentiation
GO:0010871; negative regulation of receptor biosynthetic process
GO:0010887; negative regulation of cholesterol storage
GO:0010891; negative regulation of sequestering of triglyceride
GO:0015908; fatty acid transport
GO:0030522; intracellular receptor signaling pathway
GO:0032000; positive regulation of fatty acid beta-oxidation
GO:0032091; negative regulation of protein binding
GO:0032099; negative regulation of appetite
GO:0032868; response to insulin
GO:0032922; circadian regulation of gene expression
GO:0035095; behavioral response to nicotine
GO:0042060; wound healing
GO:0042157; lipoprotein metabolic process
GO:0042752; regulation of circadian rhythm
GO:0043401; steroid hormone mediated signaling pathway
GO:0044255; cellular lipid metabolic process
GO:0045722; positive regulation of gluconeogenesis
GO:0045776; negative regulation of blood pressure
GO:0045820; negative regulation of glycolytic process
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0046321; positive regulation of fatty acid oxidation
GO:0050728; negative regulation of inflammatory response
GO:0070166; enamel mineralization
GO:0072363; regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter
GO:0072366; regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter
GO:0072369; regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter
GO:1901215; negative regulation of neuron death
GO:1902894; negative regulation of pri-miRNA transcription from RNA polymerase II promoter
GO:1903038; negative regulation of leukocyte cell-cell adhesion
GO:2000678; negative regulation of transcription regulatory region DNA binding
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
Hide GO termsFunction: GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001078; transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001103; RNA polymerase II repressing transcription factor binding
GO:0001190; transcriptional activator activity, RNA polymerase II transcription factor binding
GO:0001223; transcription coactivator binding
GO:0003677; DNA binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0003707; steroid hormone receptor activity
GO:0004879; RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0008144; drug binding
GO:0008270; zinc ion binding
GO:0008289; lipid binding
GO:0019902; phosphatase binding
GO:0019904; protein domain specific binding
GO:0031624; ubiquitin conjugating enzyme binding
GO:0032403; protein complex binding
GO:0043565; sequence-specific DNA binding
GO:0051525; NFAT protein binding
GO:0097371; MDM2/MDM4 family protein binding
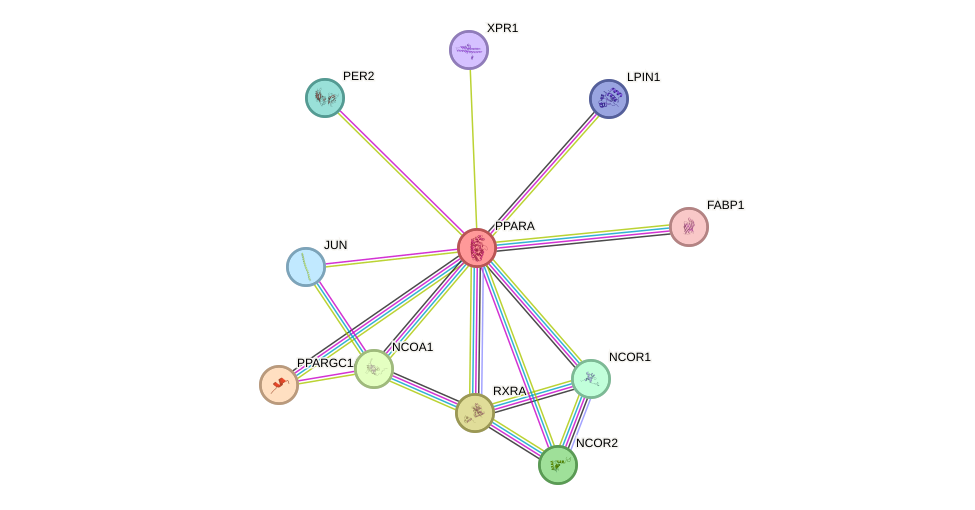
Protein interactions and network
- Protein-protein interacting partners in GenAge
- POU1F1, NCOR1, BCL2, HSP90AA1, UBE2I, EP300, GRB2, RELA, SIRT1, PPARGC1A, NCOR2
- STRING interaction network
Retrieve sequences for PPARA
Homologs in model organisms
In other databases
- GenDR gene expression
- A homolog of this gene for Mus musculus is present as Ppara
- LongevityMap
- This gene is present as PPARA