GenAge entry for E2F1 (Homo sapiens)
Gene name (HAGRID: 18)
- HGNC symbol
- E2F1
- Aliases
- RBP3; RBBP3
- Common name
- E2F transcription factor 1
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence directly linking the gene product to ageing in a cellular model system
- Description
An important regulator of the cell cycle, E2F1 may mediate apoptosis in conjunction with TP53 [85]. Clearly, E2F1 appears to be related to cancer [86]. Its role in ageing, however, is unknown.
Cytogenetic information
- Cytogenetic band
- 20q11.2
- Location
- 33,675,486 bp to 33,686,404 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000077; DNA damage checkpoint
GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0006351; transcription, DNA-templated
GO:0006355; regulation of transcription, DNA-templated
GO:0006977; DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest
GO:0007283; spermatogenesis
GO:0008630; intrinsic apoptotic signaling pathway in response to DNA damage
GO:0010628; positive regulation of gene expression
GO:0030900; forebrain development
GO:0043276; anoikis
GO:0043392; negative regulation of DNA binding
GO:0045599; negative regulation of fat cell differentiation
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0048146; positive regulation of fibroblast proliferation
GO:0048255; mRNA stabilization
GO:0070345; negative regulation of fat cell proliferation
GO:0071398; cellular response to fatty acid
GO:0071456; cellular response to hypoxia
GO:0071466; cellular response to xenobiotic stimulus
GO:0071930; negative regulation of transcription involved in G1/S transition of mitotic cell cycle
GO:0072332; intrinsic apoptotic signaling pathway by p53 class mediator
GO:1900740; positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway
GO:1990086; lens fiber cell apoptotic process
GO:1990090; cellular response to nerve growth factor stimulus
GO:2000045; regulation of G1/S transition of mitotic cell cycle
Cellular component: GO:0000790; nuclear chromatin
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005739; mitochondrion
GO:0035189; Rb-E2F complex
Hide GO termsFunction: GO:0001047; core promoter binding
GO:0003677; DNA binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0019901; protein kinase binding
GO:0043565; sequence-specific DNA binding
GO:0046983; protein dimerization activity
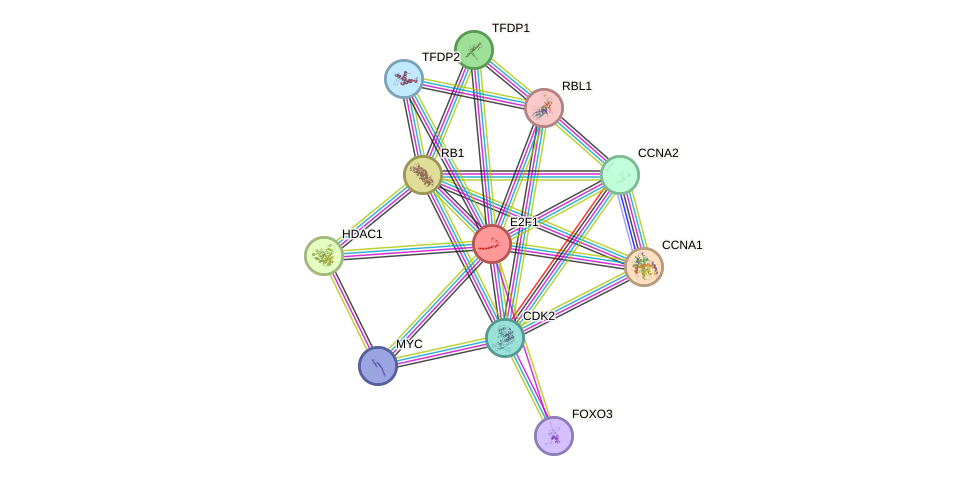
Protein interactions and network
- Protein-protein interacting partners in GenAge
- ATM, IGF1, PARP1, BRCA1, CREBBP, NFKB1, EP300, GSK3B, ERCC3, AR, RB1, SIRT1, HDAC1, SP1, CCNA2, TBP, CDK1, TFDP1, MDM2, ESR1, CDKN2A, ATR, TP53BP1, NCOR2, CDKN1A, CDK7
- STRING interaction network
Retrieve sequences for E2F1
Homologs in model organisms
- Danio rerio
- uncharacterized_H9GY
- Mus musculus
- E2f1
In other databases
- CellAge
- This gene is present as E2F1