GenAge entry for PIK3R1 (Homo sapiens)
Gene name (HAGRID: 185)
- HGNC symbol
- PIK3R1
- Aliases
- GRB1; p85-ALPHA; p85
- Common name
- phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
Together with PIK3CB, PIK3R1 is involved in insulin (INS) signalling and energy metabolism [806]. Disruption of PIK3CB in mice results in death shortly after birth and impaired B cell development [804]. Although PIK3CB has not been directly related to human ageing, PIK3CB may be associated with signalling pathways that impact on human ageing.
Cytogenetic information
- Cytogenetic band
- 5q13.1
- Location
- 68,215,756 bp to 68,301,821 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0001678; cellular glucose homeostasis
GO:0001953; negative regulation of cell-matrix adhesion
GO:0006468; protein phosphorylation
GO:0006661; phosphatidylinositol biosynthetic process
GO:0007173; epidermal growth factor receptor signaling pathway
GO:0008286; insulin receptor signaling pathway
GO:0008625; extrinsic apoptotic signaling pathway via death domain receptors
GO:0008630; intrinsic apoptotic signaling pathway in response to DNA damage
GO:0014065; phosphatidylinositol 3-kinase signaling
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0016032; viral process
GO:0030168; platelet activation
GO:0030183; B cell differentiation
GO:0030335; positive regulation of cell migration
GO:0031295; T cell costimulation
GO:0032760; positive regulation of tumor necrosis factor production
GO:0032869; cellular response to insulin stimulus
GO:0033120; positive regulation of RNA splicing
GO:0034644; cellular response to UV
GO:0034976; response to endoplasmic reticulum stress
GO:0036092; phosphatidylinositol-3-phosphate biosynthetic process
GO:0038095; Fc-epsilon receptor signaling pathway
GO:0038096; Fc-gamma receptor signaling pathway involved in phagocytosis
GO:0038128; ERBB2 signaling pathway
GO:0042993; positive regulation of transcription factor import into nucleus
GO:0043066; negative regulation of apoptotic process
GO:0043551; regulation of phosphatidylinositol 3-kinase activity
GO:0045671; negative regulation of osteoclast differentiation
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0046326; positive regulation of glucose import
GO:0046626; regulation of insulin receptor signaling pathway
GO:0046854; phosphatidylinositol phosphorylation
GO:0048009; insulin-like growth factor receptor signaling pathway
GO:0048010; vascular endothelial growth factor receptor signaling pathway
GO:0048015; phosphatidylinositol-mediated signaling
GO:0050821; protein stabilization
GO:0050852; T cell receptor signaling pathway
GO:0050900; leukocyte migration
GO:0051492; regulation of stress fiber assembly
GO:0051531; NFAT protein import into nucleus
GO:0060396; growth hormone receptor signaling pathway
GO:0090004; positive regulation of establishment of protein localization to plasma membrane
GO:1900103; positive regulation of endoplasmic reticulum unfolded protein response
GO:2001275; positive regulation of glucose import in response to insulin stimulus
Cellular component: GO:0005634; nucleus
GO:0005737; cytoplasm
GO:0005801; cis-Golgi network
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0005911; cell-cell junction
GO:0005942; phosphatidylinositol 3-kinase complex
GO:0005943; phosphatidylinositol 3-kinase complex, class IA
GO:0016020; membrane
GO:0043234; protein complex
GO:1990578; perinuclear endoplasmic reticulum membrane
Hide GO termsFunction: GO:0005068; transmembrane receptor protein tyrosine kinase adaptor activity
GO:0005158; insulin receptor binding
GO:0005159; insulin-like growth factor receptor binding
GO:0005168; neurotrophin TRKA receptor binding
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0016303; 1-phosphatidylinositol-3-kinase activity
GO:0019903; protein phosphatase binding
GO:0035014; phosphatidylinositol 3-kinase regulator activity
GO:0036312; phosphatidylinositol 3-kinase regulatory subunit binding
GO:0043125; ErbB-3 class receptor binding
GO:0043548; phosphatidylinositol 3-kinase binding
GO:0043559; insulin binding
GO:0043560; insulin receptor substrate binding
GO:0046934; phosphatidylinositol-4,5-bisphosphate 3-kinase activity
GO:0046935; 1-phosphatidylinositol-3-kinase regulator activity
GO:0046982; protein heterodimerization activity
Protein interactions and network
- Protein-protein interacting partners in GenAge
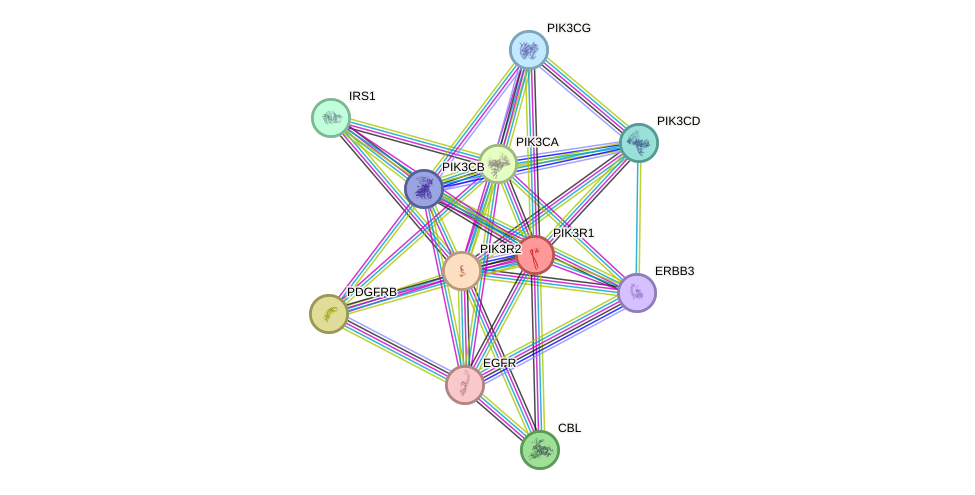
- SHC1, IGF1R, PTPN11, STAT3, HDAC3, IRS1, IRS2, PIK3CB, HRAS, EGFR, ERBB2, PDGFRB, EPOR, PLCG2, BRCA1, ABL1, EP300, HTT, GRB2, SIRT1, PTK2, FGFR1, FLT1, MAP3K5, PIK3R1, JAK2, ESR1, NFKBIA, CTNNB1, ARHGAP1, PDGFRA, PIK3CA, SQSTM1
- STRING interaction network
Retrieve sequences for PIK3R1
Homologs in model organisms
In other databases
- LongevityMap
- This gene is present as PIK3R1