GenAge entry for PSEN1 (Homo sapiens)
Gene name (HAGRID: 224)
- HGNC symbol
- PSEN1
- Aliases
- FAD; S182; PS1; AD3
- Common name
- presenilin 1
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on indirect or inconclusive evidence linking the gene product to ageing in humans or in one or more model systems
- Description
The PSEN1 gene has multiple transcriptional variants and its functions could include the cleavage of APP and notch receptor protein. PSEN1-null mice die shortly after birth [1368]. Inactivating PSEN1 in the postnatal forebrain reduced beta-amyloid generation with subtle cognitive deficits [1374]. Mutations in the human PSEN1 gene cause early-onset Alzheimer's disease [1381], though PSEN1's role in human ageing remains largely unknown.
Cytogenetic information
- Cytogenetic band
- 14q24.3
- Location
- 73,136,435 bp to 73,223,691 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000045; autophagosome assembly
GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0000186; activation of MAPKK activity
GO:0001568; blood vessel development
GO:0001708; cell fate specification
GO:0001756; somitogenesis
GO:0001764; neuron migration
GO:0001921; positive regulation of receptor recycling
GO:0001947; heart looping
GO:0002244; hematopoietic progenitor cell differentiation
GO:0002286; T cell activation involved in immune response
GO:0003407; neural retina development
GO:0006486; protein glycosylation
GO:0006509; membrane protein ectodomain proteolysis
GO:0006839; mitochondrial transport
GO:0006974; cellular response to DNA damage stimulus
GO:0006979; response to oxidative stress
GO:0007175; negative regulation of epidermal growth factor-activated receptor activity
GO:0007219; Notch signaling pathway
GO:0007220; Notch receptor processing
GO:0007613; memory
GO:0009791; post-embryonic development
GO:0015031; protein transport
GO:0015813; L-glutamate transport
GO:0015871; choline transport
GO:0016080; synaptic vesicle targeting
GO:0016337; single organismal cell-cell adhesion
GO:0016485; protein processing
GO:0021795; cerebral cortex cell migration
GO:0021870; Cajal-Retzius cell differentiation
GO:0021904; dorsal/ventral neural tube patterning
GO:0030326; embryonic limb morphogenesis
GO:0032436; positive regulation of proteasomal ubiquitin-dependent protein catabolic process
GO:0032469; endoplasmic reticulum calcium ion homeostasis
GO:0034205; beta-amyloid formation
GO:0035556; intracellular signal transduction
GO:0042325; regulation of phosphorylation
GO:0042987; amyloid precursor protein catabolic process
GO:0043011; myeloid dendritic cell differentiation
GO:0043065; positive regulation of apoptotic process
GO:0043066; negative regulation of apoptotic process
GO:0043085; positive regulation of catalytic activity
GO:0043393; regulation of protein binding
GO:0043406; positive regulation of MAP kinase activity
GO:0043524; negative regulation of neuron apoptotic process
GO:0043589; skin morphogenesis
GO:0045893; positive regulation of transcription, DNA-templated
GO:0048167; regulation of synaptic plasticity
GO:0048538; thymus development
GO:0048666; neuron development
GO:0048705; skeletal system morphogenesis
GO:0048854; brain morphogenesis
GO:0050673; epithelial cell proliferation
GO:0050771; negative regulation of axonogenesis
GO:0050820; positive regulation of coagulation
GO:0050852; T cell receptor signaling pathway
GO:0051402; neuron apoptotic process
GO:0051444; negative regulation of ubiquitin-protein transferase activity
GO:0051563; smooth endoplasmic reticulum calcium ion homeostasis
GO:0051966; regulation of synaptic transmission, glutamatergic
GO:0060070; canonical Wnt signaling pathway
GO:0060075; regulation of resting membrane potential
GO:0060999; positive regulation of dendritic spine development
GO:0070588; calcium ion transmembrane transport
GO:2000059; negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process
GO:2001234; negative regulation of apoptotic signaling pathway
Cellular component: GO:0000139; Golgi membrane
GO:0000776; kinetochore
GO:0005634; nucleus
GO:0005640; nuclear outer membrane
GO:0005739; mitochondrion
GO:0005743; mitochondrial inner membrane
GO:0005765; lysosomal membrane
GO:0005783; endoplasmic reticulum
GO:0005789; endoplasmic reticulum membrane
GO:0005790; smooth endoplasmic reticulum
GO:0005791; rough endoplasmic reticulum
GO:0005794; Golgi apparatus
GO:0005813; centrosome
GO:0005886; plasma membrane
GO:0005887; integral component of plasma membrane
GO:0009986; cell surface
GO:0016020; membrane
GO:0016021; integral component of membrane
GO:0016235; aggresome
GO:0030054; cell junction
GO:0030424; axon
GO:0030426; growth cone
GO:0031410; cytoplasmic vesicle
GO:0031594; neuromuscular junction
GO:0031965; nuclear membrane
GO:0035253; ciliary rootlet
GO:0043025; neuronal cell body
GO:0043198; dendritic shaft
GO:0045121; membrane raft
GO:0070765; gamma-secretase complex
GO:0098793; presynapse
Hide GO termsFunction: GO:0004175; endopeptidase activity
GO:0004190; aspartic-type endopeptidase activity
GO:0005262; calcium channel activity
GO:0005515; protein binding
GO:0008013; beta-catenin binding
GO:0030165; PDZ domain binding
GO:0045296; cadherin binding
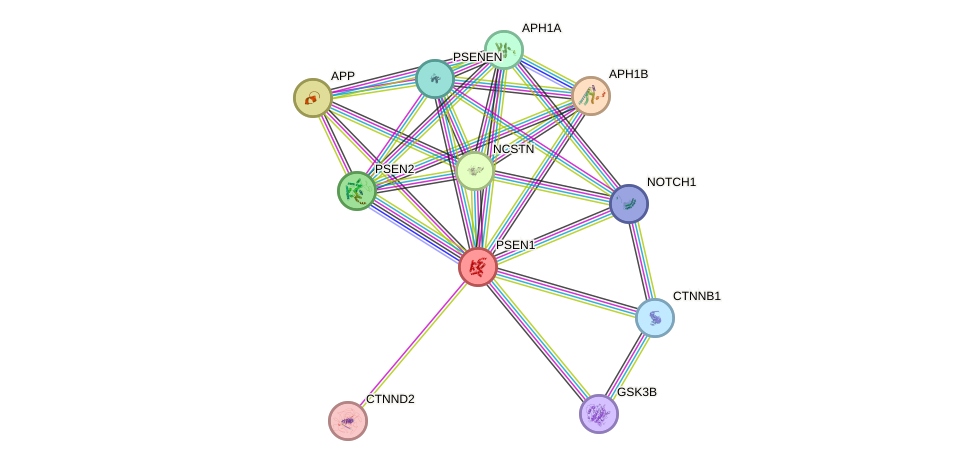
Protein interactions and network
- Protein-protein interacting partners in GenAge
- BCL2, GSK3B, APP, APOE, HMGB1, MAPT, CTNNB1, PSEN1
- STRING interaction network
Retrieve sequences for PSEN1
Homologs in model organisms
In other databases
- LongevityMap
- This gene is present as PSEN1