GenAge entry for STAT3 (Homo sapiens)
Gene name (HAGRID: 22)
- HGNC symbol
- STAT3
- Aliases
- APRF
- Common name
- signal transducer and activator of transcription 3 (acute-phase response factor)
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
Transcription factors like those of the STAT family are activated by growth factors and cytokines. Their roles in intracellular signalling pathways suggest factors such as STAT3 might be involved in ageing and/or age-related disease [266]. Expression levels of JAK-STAT signalling targets is increased with age, and STAT3 has been implicated in the regulation of self-renewal and stem cell fate in several tissues. Knockdown or pharmacological inhibition of the JAK-STAT enhances satellite stem cell division potential resulting in better muscle regeneration [3635]. STAT3 has been associated with age-related heart failure in mice [270].
Cytogenetic information
- Cytogenetic band
- 17q21.31
- Location
- 42,313,325 bp to 42,388,495 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0001659; temperature homeostasis
GO:0001754; eye photoreceptor cell differentiation
GO:0006355; regulation of transcription, DNA-templated
GO:0006357; regulation of transcription from RNA polymerase II promoter
GO:0006606; protein import into nucleus
GO:0006928; movement of cell or subcellular component
GO:0006953; acute-phase response
GO:0007165; signal transduction
GO:0007259; JAK-STAT cascade
GO:0007399; nervous system development
GO:0007568; aging
GO:0008283; cell proliferation
GO:0008284; positive regulation of cell proliferation
GO:0008285; negative regulation of cell proliferation
GO:0010628; positive regulation of gene expression
GO:0010730; negative regulation of hydrogen peroxide biosynthetic process
GO:0016032; viral process
GO:0016310; phosphorylation
GO:0019221; cytokine-mediated signaling pathway
GO:0019953; sexual reproduction
GO:0030522; intracellular receptor signaling pathway
GO:0032355; response to estradiol
GO:0032870; cellular response to hormone stimulus
GO:0033210; leptin-mediated signaling pathway
GO:0035019; somatic stem cell population maintenance
GO:0035278; miRNA mediated inhibition of translation
GO:0040014; regulation of multicellular organism growth
GO:0042493; response to drug
GO:0042517; positive regulation of tyrosine phosphorylation of Stat3 protein
GO:0042593; glucose homeostasis
GO:0042755; eating behavior
GO:0042789; mRNA transcription from RNA polymerase II promoter
GO:0043066; negative regulation of apoptotic process
GO:0044320; cellular response to leptin stimulus
GO:0044321; response to leptin
GO:0045471; response to ethanol
GO:0045747; positive regulation of Notch signaling pathway
GO:0045820; negative regulation of glycolytic process
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0046902; regulation of mitochondrial membrane permeability
GO:0048708; astrocyte differentiation
GO:0051726; regulation of cell cycle
GO:0060019; radial glial cell differentiation
GO:0060259; regulation of feeding behavior
GO:0060396; growth hormone receptor signaling pathway
GO:0060397; JAK-STAT cascade involved in growth hormone signaling pathway
GO:0070102; interleukin-6-mediated signaling pathway
GO:0071407; cellular response to organic cyclic compound
GO:0097009; energy homeostasis
GO:1901215; negative regulation of neuron death
GO:1902728; positive regulation of growth factor dependent skeletal muscle satellite cell proliferation
GO:1902895; positive regulation of pri-miRNA transcription from RNA polymerase II promoter
GO:1904685; positive regulation of metalloendopeptidase activity
GO:2000637; positive regulation of gene silencing by miRNA
GO:2000737; negative regulation of stem cell differentiation
GO:2001171; positive regulation of ATP biosynthetic process
GO:2001223; negative regulation of neuron migration
Cellular component: GO:0000790; nuclear chromatin
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005743; mitochondrial inner membrane
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0090575; RNA polymerase II transcription factor complex
Hide GO termsFunction: GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001103; RNA polymerase II repressing transcription factor binding
GO:0001228; transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding
GO:0003677; DNA binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0004871; signal transducer activity
GO:0004879; RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0019901; protein kinase binding
GO:0019903; protein phosphatase binding
GO:0031490; chromatin DNA binding
GO:0031730; CCR5 chemokine receptor binding
GO:0035259; glucocorticoid receptor binding
GO:0042802; identical protein binding
GO:0044212; transcription regulatory region DNA binding
GO:0046983; protein dimerization activity
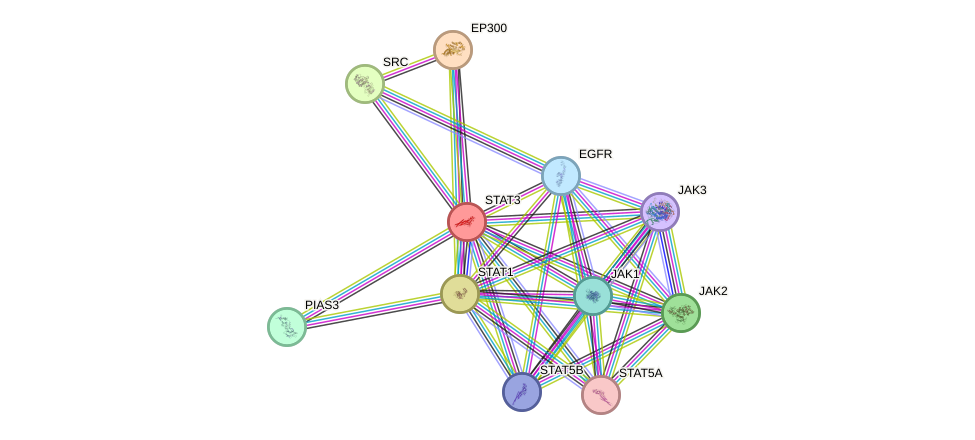
Protein interactions and network
- Protein-protein interacting partners in GenAge
- GHR, IGF1R, PTPN11, NFKB2, STAT5B, STAT3, HDAC3, PTPN1, EGFR, ERBB2, PDGFRB, PRKCD, RET, CREBBP, HIF1A, HSP90AA1, NR3C1, NFKB1, CEBPB, EP300, PML, AR, RB1, RELA, SIRT1, HDAC1, MAPK8, PTK2, FGFR1, SP1, JUN, HESX1, PIK3R1, APEX1, SIN3A, HDAC2, JAK2, ESR1, MTOR, HIC1, CDKN1A, PDGFRA
- STRING interaction network