GenAge entry for PTPN11 (Homo sapiens)
Gene name (HAGRID: 19)
- HGNC symbol
- PTPN11
- Aliases
- BPTP3; SH-PTP2; SHP-2; PTP2C; SHP2; NS1
- Common name
- protein tyrosine phosphatase, non-receptor type 11
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
PTPN11 is a signalling protein that plays a role in the signal transduction of many receptors that have been associated with ageing, such as GHR [1559]. Mutations in PTPN11 cause Noonan syndrome, which is often characterized by growth impairment and a disturbance of GH1 secretion and IGF1 signalling, even though this disease is probably due to a gain-of-function mutation [257]. It is unknown whether PTPN11 plays a role in human ageing, but it may play a role in its signalling cascade.
Cytogenetic information
- Cytogenetic band
- 12q24
- Location
- 112,418,732 bp to 112,509,913 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000077; DNA damage checkpoint
GO:0000187; activation of MAPK activity
GO:0006641; triglyceride metabolic process
GO:0007173; epidermal growth factor receptor signaling pathway
GO:0007229; integrin-mediated signaling pathway
GO:0007409; axonogenesis
GO:0007420; brain development
GO:0007507; heart development
GO:0008543; fibroblast growth factor receptor signaling pathway
GO:0009755; hormone-mediated signaling pathway
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0021697; cerebellar cortex formation
GO:0030168; platelet activation
GO:0030220; platelet formation
GO:0031295; T cell costimulation
GO:0032528; microvillus organization
GO:0033277; abortive mitotic cell cycle
GO:0033628; regulation of cell adhesion mediated by integrin
GO:0033629; negative regulation of cell adhesion mediated by integrin
GO:0035264; multicellular organism growth
GO:0035265; organ growth
GO:0035335; peptidyl-tyrosine dephosphorylation
GO:0035855; megakaryocyte development
GO:0036092; phosphatidylinositol-3-phosphate biosynthetic process
GO:0036302; atrioventricular canal development
GO:0038127; ERBB signaling pathway
GO:0040014; regulation of multicellular organism growth
GO:0042445; hormone metabolic process
GO:0042593; glucose homeostasis
GO:0043254; regulation of protein complex assembly
GO:0045931; positive regulation of mitotic cell cycle
GO:0046676; negative regulation of insulin secretion
GO:0046825; regulation of protein export from nucleus
GO:0046854; phosphatidylinositol phosphorylation
GO:0046887; positive regulation of hormone secretion
GO:0048008; platelet-derived growth factor receptor signaling pathway
GO:0048011; neurotrophin TRK receptor signaling pathway
GO:0048013; ephrin receptor signaling pathway
GO:0048015; phosphatidylinositol-mediated signaling
GO:0048609; multicellular organismal reproductive process
GO:0048806; genitalia development
GO:0048839; inner ear development
GO:0048873; homeostasis of number of cells within a tissue
GO:0050900; leukocyte migration
GO:0051463; negative regulation of cortisol secretion
GO:0060020; Bergmann glial cell differentiation
GO:0060125; negative regulation of growth hormone secretion
GO:0060325; face morphogenesis
GO:0060338; regulation of type I interferon-mediated signaling pathway
GO:0061582; intestinal epithelial cell migration
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:2001275; positive regulation of glucose import in response to insulin stimulus
Cellular component: GO:0005634; nucleus
GO:0005737; cytoplasm
GO:0005739; mitochondrion
GO:0005829; cytosol
GO:0043234; protein complex
Hide GO termsFunction: GO:0004721; phosphoprotein phosphatase activity
GO:0004725; protein tyrosine phosphatase activity
GO:0004726; non-membrane spanning protein tyrosine phosphatase activity
GO:0005070; SH3/SH2 adaptor activity
GO:0005158; insulin receptor binding
GO:0005515; protein binding
GO:0016303; 1-phosphatidylinositol-3-kinase activity
GO:0019904; protein domain specific binding
GO:0030971; receptor tyrosine kinase binding
GO:0031748; D1 dopamine receptor binding
GO:0043274; phospholipase binding
GO:0043560; insulin receptor substrate binding
GO:0046934; phosphatidylinositol-4,5-bisphosphate 3-kinase activity
GO:0050839; cell adhesion molecule binding
GO:0051428; peptide hormone receptor binding
Protein interactions and network
- Protein-protein interacting partners in GenAge
- GHR, IGF1R, PTPN11, STAT5B, STAT3, STAT5A, IRS1, IRS2, EGFR, ERBB2, INSR, PDGFRB, EPOR, PLCG2, GSK3B, GRB2, PTK2B, PTK2, MAPK14, FLT1, MAP3K5, PIK3R1, JAK2, CTNNB1
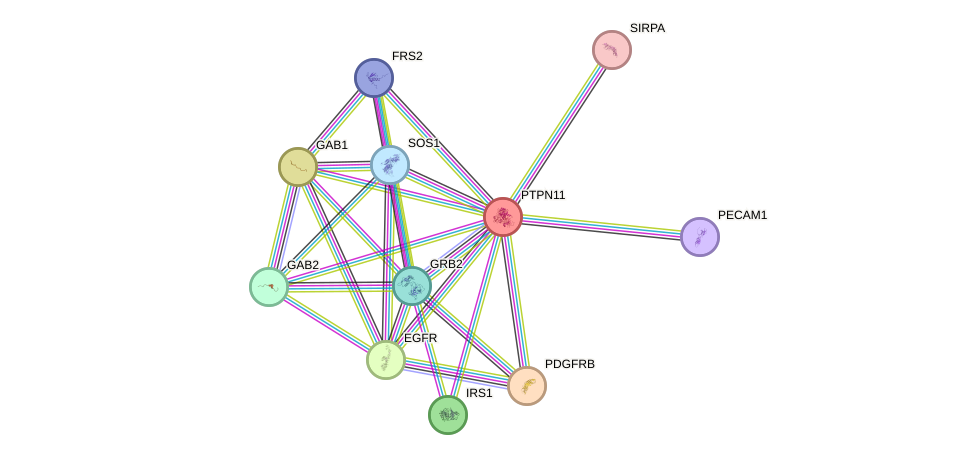
- STRING interaction network
Retrieve sequences for PTPN11
Homologs in model organisms
- Caenorhabditis elegans
- CELE_ZK616.8
- Danio rerio
- ptpn11a
- Drosophila melanogaster
- csw
- Mus musculus
- Ptpn11
- Rattus norvegicus
- Ptpn11
- Schizosaccharomyces pombe
- pyp3
In other databases
- LongevityMap
- This gene is present as PTPN11