GenAge entry for GRB2 (Homo sapiens)
Gene name (HAGRID: 122)
- HGNC symbol
- GRB2
- Aliases
- NCKAP2
- Common name
- growth factor receptor-bound protein 2
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence showing that the gene product to be a target of genes previously linked to ageing
- Description
An important player in growth signalling pathways, GRB2's functions are not yet well understood. Analyses of mice with a null mutation in GRB2 suggest that GRB2 is required for embryonic development and malignant transformation [620]. Due to its role as a signal transducer of many pathways, including pathways and genes previously related to ageing such as INS/IGF1 signalling and SHC1 [1107], it is possible GRB2 plays some role in ageing.
Cytogenetic information
- Cytogenetic band
- 17q24-q25
- Location
- 75,318,076 bp to 75,405,708 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000165; MAPK cascade
GO:0007173; epidermal growth factor receptor signaling pathway
GO:0007265; Ras protein signal transduction
GO:0007267; cell-cell signaling
GO:0007411; axon guidance
GO:0007568; aging
GO:0008286; insulin receptor signaling pathway
GO:0008543; fibroblast growth factor receptor signaling pathway
GO:0009967; positive regulation of signal transduction
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0016032; viral process
GO:0030838; positive regulation of actin filament polymerization
GO:0031295; T cell costimulation
GO:0031623; receptor internalization
GO:0036092; phosphatidylinositol-3-phosphate biosynthetic process
GO:0038095; Fc-epsilon receptor signaling pathway
GO:0038096; Fc-gamma receptor signaling pathway involved in phagocytosis
GO:0038128; ERBB2 signaling pathway
GO:0042059; negative regulation of epidermal growth factor receptor signaling pathway
GO:0042770; signal transduction in response to DNA damage
GO:0043408; regulation of MAPK cascade
GO:0043547; positive regulation of GTPase activity
GO:0046854; phosphatidylinositol phosphorylation
GO:0048015; phosphatidylinositol-mediated signaling
GO:0048646; anatomical structure formation involved in morphogenesis
GO:0050900; leukocyte migration
GO:0051291; protein heterooligomerization
GO:0060670; branching involved in labyrinthine layer morphogenesis
GO:0071479; cellular response to ionizing radiation
GO:2000379; positive regulation of reactive oxygen species metabolic process
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005730; nucleolus
GO:0005737; cytoplasm
GO:0005768; endosome
GO:0005794; Golgi apparatus
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0005911; cell-cell junction
GO:0008180; COP9 signalosome
GO:0012506; vesicle membrane
GO:0070062; extracellular exosome
GO:0070436; Grb2-EGFR complex
Hide GO termsFunction: GO:0001784; phosphotyrosine binding
GO:0005070; SH3/SH2 adaptor activity
GO:0005088; Ras guanyl-nucleotide exchange factor activity
GO:0005154; epidermal growth factor receptor binding
GO:0005168; neurotrophin TRKA receptor binding
GO:0005515; protein binding
GO:0016303; 1-phosphatidylinositol-3-kinase activity
GO:0017124; SH3 domain binding
GO:0019901; protein kinase binding
GO:0019903; protein phosphatase binding
GO:0042802; identical protein binding
GO:0043560; insulin receptor substrate binding
GO:0044822; poly(A) RNA binding
GO:0046875; ephrin receptor binding
GO:0046934; phosphatidylinositol-4,5-bisphosphate 3-kinase activity
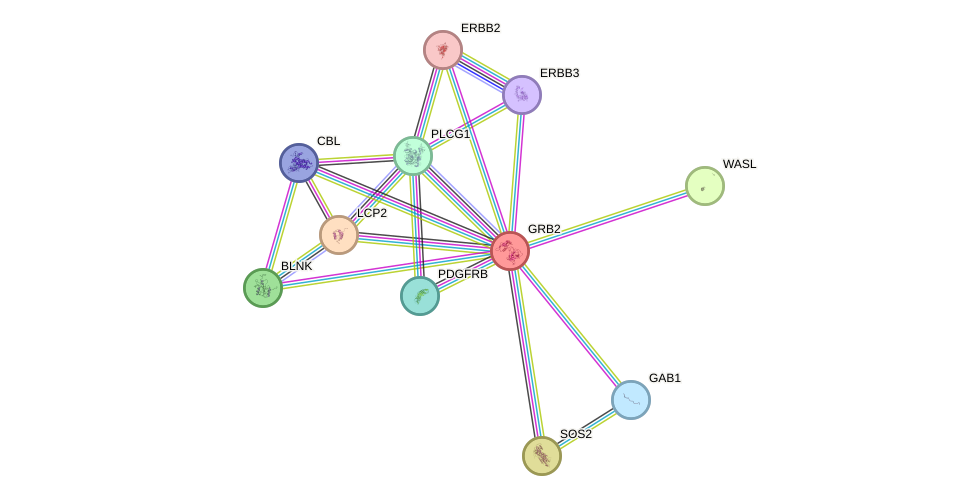
Protein interactions and network
- Protein-protein interacting partners in GenAge
- GHR, SHC1, PTPN11, IRS1, PTPN1, IRS2, PIK3CB, EGFR, ERBB2, INSR, PDGFRB, EPOR, PPARA, RET, VCP, HSP90AA1, ABL1, EP300, HTT, RB1, GRB2, A2M, HSPA9, HSPD1, HSPA1B, YWHAZ, PTK2B, PTK2, MAPK9, PIK3R1, HSPA8, JAK2, CLU, PPP1CA, DBN1, CDC42, EPS8, PDGFRA, PIK3CA
- STRING interaction network
Retrieve sequences for GRB2
Homologs in model organisms
In other databases
- LongevityMap
- This gene is present as GRB2