GenAge entry for RB1 (Homo sapiens)
Gene name (HAGRID: 120)
- HGNC symbol
- RB1
- Aliases
- RB; PPP1R130; OSRC
- Common name
- retinoblastoma 1
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence directly linking the gene product to ageing in a cellular model system
- Description
RB1 is a tumour suppressor that acts as a transcriptional regulator to control cell cycle progression. Mice with disrupted RB1 are not viable [605]. Mutations in the human RB1 gene have been associated with cancer [991]. Clearly, RB1 plays a role in cellular senescence and oncogenesis [95], but it is unknown whether it influences human ageing.
Cytogenetic information
- Cytogenetic band
- 13q14.2
- Location
- 48,303,747 bp to 48,481,890 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000075; cell cycle checkpoint
GO:0000082; G1/S transition of mitotic cell cycle
GO:0000083; regulation of transcription involved in G1/S transition of mitotic cell cycle
GO:0001558; regulation of cell growth
GO:0001894; tissue homeostasis
GO:0006338; chromatin remodeling
GO:0006351; transcription, DNA-templated
GO:0006469; negative regulation of protein kinase activity
GO:0007050; cell cycle arrest
GO:0007070; negative regulation of transcription from RNA polymerase II promoter during mitotic cell cycle
GO:0007093; mitotic cell cycle checkpoint
GO:0007265; Ras protein signal transduction
GO:0007346; regulation of mitotic cell cycle
GO:0010629; negative regulation of gene expression
GO:0016032; viral process
GO:0016569; covalent chromatin modification
GO:0030521; androgen receptor signaling pathway
GO:0031134; sister chromatid biorientation
GO:0031175; neuron projection development
GO:0034088; maintenance of mitotic sister chromatid cohesion
GO:0034349; glial cell apoptotic process
GO:0035914; skeletal muscle cell differentiation
GO:0042551; neuron maturation
GO:0043353; enucleate erythrocyte differentiation
GO:0043433; negative regulation of sequence-specific DNA binding transcription factor activity
GO:0043550; regulation of lipid kinase activity
GO:0045445; myoblast differentiation
GO:0045651; positive regulation of macrophage differentiation
GO:0045842; positive regulation of mitotic metaphase/anaphase transition
GO:0045879; negative regulation of smoothened signaling pathway
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0048565; digestive tract development
GO:0048667; cell morphogenesis involved in neuron differentiation
GO:0050680; negative regulation of epithelial cell proliferation
GO:0051146; striated muscle cell differentiation
GO:0051301; cell division
GO:0051402; neuron apoptotic process
GO:0071459; protein localization to chromosome, centromeric region
GO:0071466; cellular response to xenobiotic stimulus
GO:0071922; regulation of cohesin loading
GO:0071930; negative regulation of transcription involved in G1/S transition of mitotic cell cycle
GO:0090230; regulation of centromere complex assembly
GO:0097284; hepatocyte apoptotic process
GO:2000134; negative regulation of G1/S transition of mitotic cell cycle
GO:2000679; positive regulation of transcription regulatory region DNA binding
Cellular component: GO:0000785; chromatin
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005819; spindle
GO:0008024; cyclin/CDK positive transcription elongation factor complex
GO:0016514; SWI/SNF complex
GO:0016605; PML body
GO:0035189; Rb-E2F complex
Hide GO termsFunction: GO:0001047; core promoter binding
GO:0001102; RNA polymerase II activating transcription factor binding
GO:0003677; DNA binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0003713; transcription coactivator activity
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0019900; kinase binding
GO:0031625; ubiquitin protein ligase binding
GO:0042802; identical protein binding
GO:0050681; androgen receptor binding
GO:0051219; phosphoprotein binding
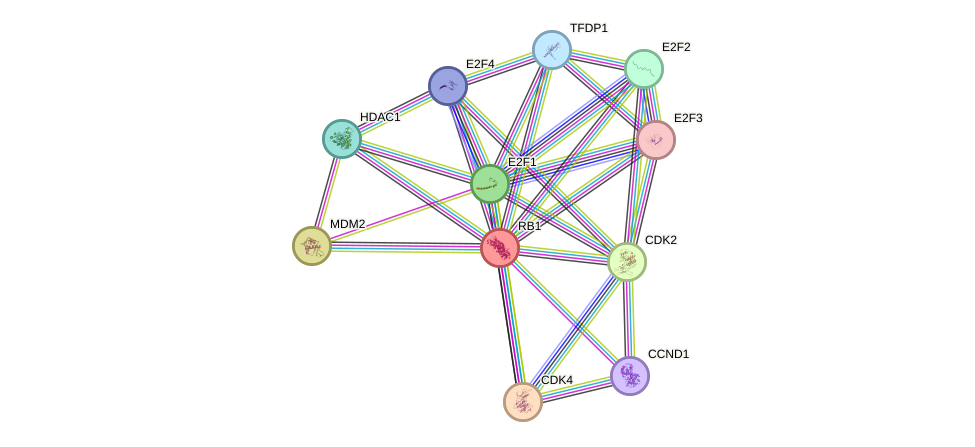
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, LMNA, E2F1, STAT3, HDAC3, INS, MYC, FOS, TCF3, BRCA1, HIF1A, ABL1, TOP2A, UBE2I, CEBPA, CEBPB, EP300, PML, AR, FAS, GRB2, FOXO3, FOXO1, FOXM1, SIRT1, HDAC1, MAPK8, MAPK14, SP1, JUN, HMGB1, CCNA2, TAF1, TFAP2A, ATF2, TBP, HBP1, CDK1, TFDP1, POLA1, HDAC2, MDM2, PPP1CA, CHEK2, PPARG, TP73, CDKN1A, TRAP1
- STRING interaction network
Retrieve sequences for RB1
Homologs in model organisms
In other databases
- CellAge
- This gene is present as RB1