GenAge entry for RELA (Homo sapiens)
Gene name (HAGRID: 143)
- HGNC symbol
- RELA
- Aliases
- p65; NFKB3
- Common name
- v-rel avian reticuloendotheliosis viral oncogene homolog A
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
Also known as NFKB3, RELA is a component of the NF-kappa-B complex, involved in oxidative pathways and a myriad of other processes. Its role in ageing is unclear, but the NF-kappa-B complex appears to be important in age-related changes in inflammation and maybe in other age-related processes too [260]. In addition, age-related changes in NF-kappa-B have been reported in numerous tissues [261]. Mice without RELA die at embryonic stages [712].
Cytogenetic information
- Cytogenetic band
- 11q13
- Location
- 65,653,596 bp to 65,662,972 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0001889; liver development
GO:0001942; hair follicle development
GO:0002223; stimulatory C-type lectin receptor signaling pathway
GO:0006117; acetaldehyde metabolic process
GO:0006366; transcription from RNA polymerase II promoter
GO:0006954; inflammatory response
GO:0006968; cellular defense response
GO:0007568; aging
GO:0008284; positive regulation of cell proliferation
GO:0009887; animal organ morphogenesis
GO:0010033; response to organic substance
GO:0010224; response to UV-B
GO:0014040; positive regulation of Schwann cell differentiation
GO:0019221; cytokine-mediated signaling pathway
GO:0031293; membrane protein intracellular domain proteolysis
GO:0032332; positive regulation of chondrocyte differentiation
GO:0032481; positive regulation of type I interferon production
GO:0032495; response to muramyl dipeptide
GO:0032570; response to progesterone
GO:0032868; response to insulin
GO:0033590; response to cobalamin
GO:0035729; cellular response to hepatocyte growth factor stimulus
GO:0035994; response to muscle stretch
GO:0038095; Fc-epsilon receptor signaling pathway
GO:0042177; negative regulation of protein catabolic process
GO:0042493; response to drug
GO:0043066; negative regulation of apoptotic process
GO:0043123; positive regulation of I-kappaB kinase/NF-kappaB signaling
GO:0043200; response to amino acid
GO:0043278; response to morphine
GO:0045084; positive regulation of interleukin-12 biosynthetic process
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0046627; negative regulation of insulin receptor signaling pathway
GO:0050727; regulation of inflammatory response
GO:0050852; T cell receptor signaling pathway
GO:0051092; positive regulation of NF-kappaB transcription factor activity
GO:0051591; response to cAMP
GO:0051607; defense response to virus
GO:0070301; cellular response to hydrogen peroxide
GO:0070431; nucleotide-binding oligomerization domain containing 2 signaling pathway
GO:0070555; response to interleukin-1
GO:0071222; cellular response to lipopolysaccharide
GO:0071316; cellular response to nicotine
GO:0071347; cellular response to interleukin-1
GO:0071354; cellular response to interleukin-6
GO:0071356; cellular response to tumor necrosis factor
GO:0071375; cellular response to peptide hormone stimulus
GO:1901223; negative regulation of NIK/NF-kappaB signaling
GO:1902895; positive regulation of pri-miRNA transcription from RNA polymerase II promoter
GO:2000630; positive regulation of miRNA metabolic process
GO:2001237; negative regulation of extrinsic apoptotic signaling pathway
Cellular component: GO:0000790; nuclear chromatin
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005667; transcription factor complex
GO:0005737; cytoplasm
GO:0005829; cytosol
GO:0033256; I-kappaB/NF-kappaB complex
GO:0071159; NF-kappaB complex
Hide GO termsFunction: GO:0000977; RNA polymerase II regulatory region sequence-specific DNA binding
GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0000980; RNA polymerase II distal enhancer sequence-specific DNA binding
GO:0000983; transcription factor activity, RNA polymerase II core promoter sequence-specific
GO:0001046; core promoter sequence-specific DNA binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001078; transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001205; transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding
GO:0003677; DNA binding
GO:0003682; chromatin binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0003705; transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0019901; protein kinase binding
GO:0031490; chromatin DNA binding
GO:0031625; ubiquitin protein ligase binding
GO:0032403; protein complex binding
GO:0033613; activating transcription factor binding
GO:0042301; phosphate ion binding
GO:0042802; identical protein binding
GO:0042803; protein homodimerization activity
GO:0042805; actinin binding
GO:0042826; histone deacetylase binding
GO:0044212; transcription regulatory region DNA binding
GO:0046982; protein heterodimerization activity
GO:0047485; protein N-terminus binding
GO:0051059; NF-kappaB binding
GO:0070491; repressing transcription factor binding
GO:0071532; ankyrin repeat binding
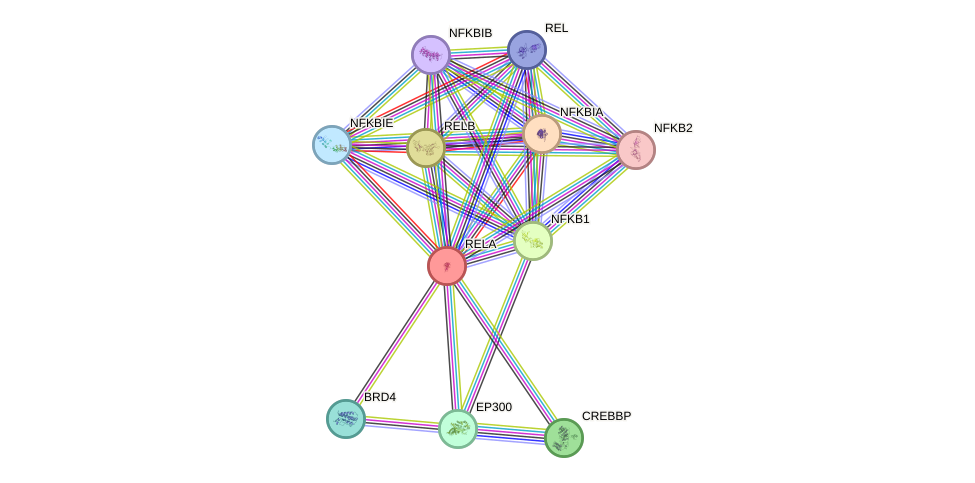
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, TERT, ATM, NFKB2, STAT3, HDAC3, MYC, FOS, PPARA, PARP1, BRCA1, PIN1, CREBBP, HIF1A, NR3C1, EGR1, NFKB1, CEBPB, EP300, PML, GSK3B, PRKDC, AR, RELA, SIRT1, HDAC1, HSPA1A, MAPK14, SP1, JUN, HMGB1, TAF1, TFAP2A, TBP, HSPA8, HDAC2, MDM2, ESR1, NFKBIA, CDKN2A, TP63, SIRT6, STUB1, PPARGC1A, PPARG, NCOR2, IKBKB
- STRING interaction network