GenAge entry for AKT1 (Homo sapiens)
Gene name (HAGRID: 35)
- HGNC symbol
- AKT1
- Aliases
- RAC; PKB; PRKBA; AKT
- Common name
- v-akt murine thymoma viral oncogene homolog 1
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence directly linking the gene product to ageing in a non-mammalian animal model
- Description
AKT1 is a protein kinase that is also considered an oncogene. It appears to mediate insulin's (INS) actions and be involved in the signal transduction of growth factors such as IGF1. AKT1 also impacts on cell survival and development, and it activates NFKB1 [333]. AKT1 null mice are smaller but viable and more susceptible to genotoxic stress and apoptosis [327]. Haploinsufficient mice have a lower body weight and a slightly longer lifespan (8% for males and 15% for females) [3259].
In roundworms, two AKT1 homologues play a role in insulin-like signalling and influence dauer formation [948]. Knockdown of akt-1 in roundworms extended lifespan [3259]. AKT1 also appears to play a role in cellular senescence [1660]. Some evidence suggests AKT1 may play a role in ageing through its impact on energy metabolism [823], but its role, if any, on human ageing remains elusive.
Cytogenetic information
- Cytogenetic band
- 14q32.32
- Location
- 104,769,350 bp to 104,793,601 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0000060; protein import into nucleus, translocation
GO:0001649; osteoblast differentiation
GO:0001893; maternal placenta development
GO:0001934; positive regulation of protein phosphorylation
GO:0001938; positive regulation of endothelial cell proliferation
GO:0005978; glycogen biosynthetic process
GO:0005979; regulation of glycogen biosynthetic process
GO:0006006; glucose metabolic process
GO:0006412; translation
GO:0006417; regulation of translation
GO:0006464; cellular protein modification process
GO:0006468; protein phosphorylation
GO:0006469; negative regulation of protein kinase activity
GO:0006809; nitric oxide biosynthetic process
GO:0006924; activation-induced cell death of T cells
GO:0006954; inflammatory response
GO:0006974; cellular response to DNA damage stimulus
GO:0006979; response to oxidative stress
GO:0007165; signal transduction
GO:0007186; G-protein coupled receptor signaling pathway
GO:0007281; germ cell development
GO:0007568; aging
GO:0008283; cell proliferation
GO:0008286; insulin receptor signaling pathway
GO:0008637; apoptotic mitochondrial changes
GO:0009408; response to heat
GO:0010507; negative regulation of autophagy
GO:0010629; negative regulation of gene expression
GO:0010748; negative regulation of plasma membrane long-chain fatty acid transport
GO:0010763; positive regulation of fibroblast migration
GO:0010765; positive regulation of sodium ion transport
GO:0010907; positive regulation of glucose metabolic process
GO:0010951; negative regulation of endopeptidase activity
GO:0010975; regulation of neuron projection development
GO:0014066; regulation of phosphatidylinositol 3-kinase signaling
GO:0015758; glucose transport
GO:0016310; phosphorylation
GO:0016567; protein ubiquitination
GO:0018105; peptidyl-serine phosphorylation
GO:0018107; peptidyl-threonine phosphorylation
GO:0021510; spinal cord development
GO:0030030; cell projection organization
GO:0030154; cell differentiation
GO:0030163; protein catabolic process
GO:0030168; platelet activation
GO:0030212; hyaluronan metabolic process
GO:0030307; positive regulation of cell growth
GO:0030334; regulation of cell migration
GO:0031018; endocrine pancreas development
GO:0031295; T cell costimulation
GO:0031641; regulation of myelination
GO:0031659; positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle
GO:0031663; lipopolysaccharide-mediated signaling pathway
GO:0031999; negative regulation of fatty acid beta-oxidation
GO:0032079; positive regulation of endodeoxyribonuclease activity
GO:0032094; response to food
GO:0032270; positive regulation of cellular protein metabolic process
GO:0032287; peripheral nervous system myelin maintenance
GO:0032436; positive regulation of proteasomal ubiquitin-dependent protein catabolic process
GO:0032869; cellular response to insulin stimulus
GO:0033138; positive regulation of peptidyl-serine phosphorylation
GO:0034405; response to fluid shear stress
GO:0035556; intracellular signal transduction
GO:0035655; interleukin-18-mediated signaling pathway
GO:0035924; cellular response to vascular endothelial growth factor stimulus
GO:0038128; ERBB2 signaling pathway
GO:0042593; glucose homeostasis
GO:0043065; positive regulation of apoptotic process
GO:0043066; negative regulation of apoptotic process
GO:0043154; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process
GO:0043488; regulation of mRNA stability
GO:0043491; protein kinase B signaling
GO:0043536; positive regulation of blood vessel endothelial cell migration
GO:0045429; positive regulation of nitric oxide biosynthetic process
GO:0045600; positive regulation of fat cell differentiation
GO:0045725; positive regulation of glycogen biosynthetic process
GO:0045742; positive regulation of epidermal growth factor receptor signaling pathway
GO:0045792; negative regulation of cell size
GO:0045861; negative regulation of proteolysis
GO:0045907; positive regulation of vasoconstriction
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0046326; positive regulation of glucose import
GO:0046329; negative regulation of JNK cascade
GO:0046622; positive regulation of organ growth
GO:0046777; protein autophosphorylation
GO:0046889; positive regulation of lipid biosynthetic process
GO:0048009; insulin-like growth factor receptor signaling pathway
GO:0048015; phosphatidylinositol-mediated signaling
GO:0048661; positive regulation of smooth muscle cell proliferation
GO:0050999; regulation of nitric-oxide synthase activity
GO:0051000; positive regulation of nitric-oxide synthase activity
GO:0051091; positive regulation of sequence-specific DNA binding transcription factor activity
GO:0051146; striated muscle cell differentiation
GO:0060416; response to growth hormone
GO:0060644; mammary gland epithelial cell differentiation
GO:0060709; glycogen cell differentiation involved in embryonic placenta development
GO:0060716; labyrinthine layer blood vessel development
GO:0070141; response to UV-A
GO:0071260; cellular response to mechanical stimulus
GO:0071364; cellular response to epidermal growth factor stimulus
GO:0071380; cellular response to prostaglandin E stimulus
GO:0071407; cellular response to organic cyclic compound
GO:0071456; cellular response to hypoxia
GO:0072655; establishment of protein localization to mitochondrion
GO:0072656; maintenance of protein location in mitochondrion
GO:0090004; positive regulation of establishment of protein localization to plasma membrane
GO:0090201; negative regulation of release of cytochrome c from mitochondria
GO:0097011; cellular response to granulocyte macrophage colony-stimulating factor stimulus
GO:0097194; execution phase of apoptosis
GO:0099565; chemical synaptic transmission, postsynaptic
GO:0100002; negative regulation of protein kinase activity by protein phosphorylation
GO:1900182; positive regulation of protein localization to nucleus
GO:1901215; negative regulation of neuron death
GO:1901796; regulation of signal transduction by p53 class mediator
GO:1901976; regulation of cell cycle checkpoint
GO:1902176; negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway
GO:1990090; cellular response to nerve growth factor stimulus
GO:1990418; response to insulin-like growth factor stimulus
GO:2001240; negative regulation of extrinsic apoptotic signaling pathway in absence of ligand
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005739; mitochondrion
GO:0005819; spindle
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0005911; cell-cell junction
GO:0015630; microtubule cytoskeleton
GO:0031982; vesicle
GO:0036064; ciliary basal body
GO:0043234; protein complex
GO:0098794; postsynapse
Hide GO termsFunction: GO:0004672; protein kinase activity
GO:0004674; protein serine/threonine kinase activity
GO:0004712; protein serine/threonine/tyrosine kinase activity
GO:0005080; protein kinase C binding
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0005547; phosphatidylinositol-3,4,5-trisphosphate binding
GO:0016301; kinase activity
GO:0019899; enzyme binding
GO:0030235; nitric-oxide synthase regulator activity
GO:0032794; GTPase activating protein binding
GO:0042802; identical protein binding
GO:0043325; phosphatidylinositol-3,4-bisphosphate binding
GO:0051721; protein phosphatase 2A binding
GO:0071889; 14-3-3 protein binding
Protein interactions and network
- Protein-protein interacting partners in GenAge
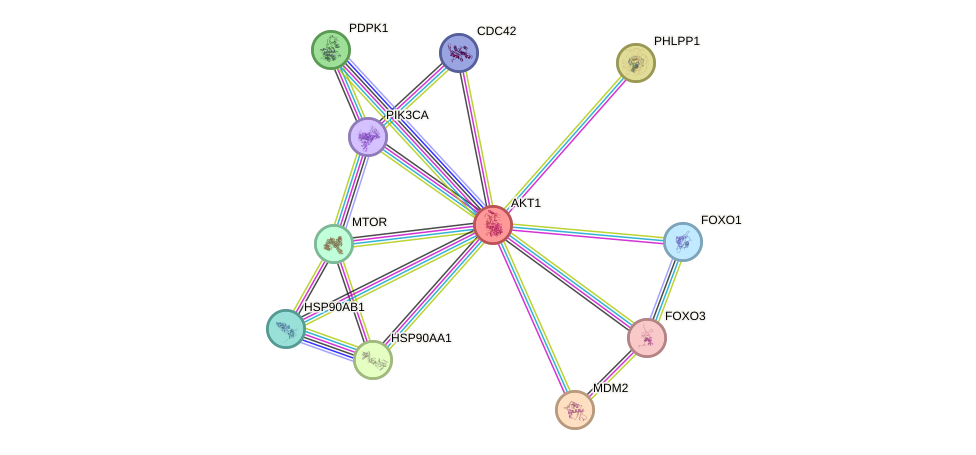
- TERT, IRS1, AKT1, PIK3CB, EGFR, HDAC3, BRCA1, PTEN, CREBBP, HSP90AA1, EP300, GSK3B, PRKDC, AR, HSPA8, FOXO3, FOXO1, UCHL1, APP, SIRT1, MAPK8, YWHAZ, MAPK14, MAPK9, MAP3K5, FOXO4, CREB1, HSPA9, MXD1, MDM2, VCP, ESR1, MTOR, CTNNB1, PPP1CA, MAPT, SIRT6, PDPK1, STUB1, NCOR2, NFE2L2, CDKN1A, PIK3CA, GSK3A, IKBKB, RICTOR
- STRING interaction network
Retrieve sequences for AKT1
Homologs in model organisms
In other databases
- GenAge model organism genes
- A homolog of this gene for Mus musculus is present as Akt1
- LongevityMap
- This gene is present as AKT1
- CellAge
- This gene is present as AKT1