GenAge entry for PRKDC (Homo sapiens)
Gene name (HAGRID: 106)
- HGNC symbol
- PRKDC
- Aliases
- DNPK1; p350; DNAPK; XRCC7; DNA-PKcs; HYRC; HYRC1
- Common name
- protein kinase, DNA-activated, catalytic polypeptide
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
PRKDC is involved in DNA repair and may modulate transcription [554]. Evidence also suggests PRKDC may be involved in telomere biology [557]. DNA breaks activate PRKDC in mouse skeletal muscle, which then goes on to phosphorylate HSP90AA1, decreasing its function as a chaperone. This suppresses mitochondrial function, energy metabolism, and physical fitness. Therefore, accumulation of DNA breaks in ageing may be one of the drivers of metabolic and fitness decline due to increased PRKDC activity [4507].
Because PRKDC has been associated with many pathways with genes likely to be involved in human ageing, such as WRN [558], it is possible that PRKDC itself is involved in human ageing, though further research is necessary to investigate this hypothesis.
Cytogenetic information
- Cytogenetic band
- 8q11
- Location
- 47,773,108 bp to 47,960,183 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0001756; somitogenesis
GO:0001933; negative regulation of protein phosphorylation
GO:0002326; B cell lineage commitment
GO:0002328; pro-B cell differentiation
GO:0002360; T cell lineage commitment
GO:0002638; negative regulation of immunoglobulin production
GO:0002684; positive regulation of immune system process
GO:0006302; double-strand break repair
GO:0006303; double-strand break repair via nonhomologous end joining
GO:0006464; cellular protein modification process
GO:0007420; brain development
GO:0007507; heart development
GO:0008283; cell proliferation
GO:0008630; intrinsic apoptotic signaling pathway in response to DNA damage
GO:0010332; response to gamma radiation
GO:0014823; response to activity
GO:0016233; telomere capping
GO:0018105; peptidyl-serine phosphorylation
GO:0031648; protein destabilization
GO:0032481; positive regulation of type I interferon production
GO:0032869; cellular response to insulin stimulus
GO:0033077; T cell differentiation in thymus
GO:0033152; immunoglobulin V(D)J recombination
GO:0033153; T cell receptor V(D)J recombination
GO:0035234; ectopic germ cell programmed cell death
GO:0042752; regulation of circadian rhythm
GO:0043065; positive regulation of apoptotic process
GO:0043066; negative regulation of apoptotic process
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0048146; positive regulation of fibroblast proliferation
GO:0048511; rhythmic process
GO:0048536; spleen development
GO:0048538; thymus development
GO:0048639; positive regulation of developmental growth
GO:0048660; regulation of smooth muscle cell proliferation
GO:0072431; signal transduction involved in mitotic G1 DNA damage checkpoint
GO:0097681; double-strand break repair via alternative nonhomologous end joining
GO:2000773; negative regulation of cellular senescence
GO:2001229; negative regulation of response to gamma radiation
Cellular component: GO:0000784; nuclear chromosome, telomeric region
GO:0005654; nucleoplasm
GO:0005667; transcription factor complex
GO:0005730; nucleolus
GO:0005829; cytosol
GO:0005958; DNA-dependent protein kinase-DNA ligase 4 complex
GO:0016020; membrane
GO:0031012; extracellular matrix
GO:0070419; nonhomologous end joining complex
Hide GO termsFunction: GO:0003690; double-stranded DNA binding
GO:0004672; protein kinase activity
GO:0004674; protein serine/threonine kinase activity
GO:0004677; DNA-dependent protein kinase activity
GO:0005515; protein binding
GO:0005524; ATP binding
GO:0008134; transcription factor binding
GO:0019899; enzyme binding
GO:0044822; poly(A) RNA binding
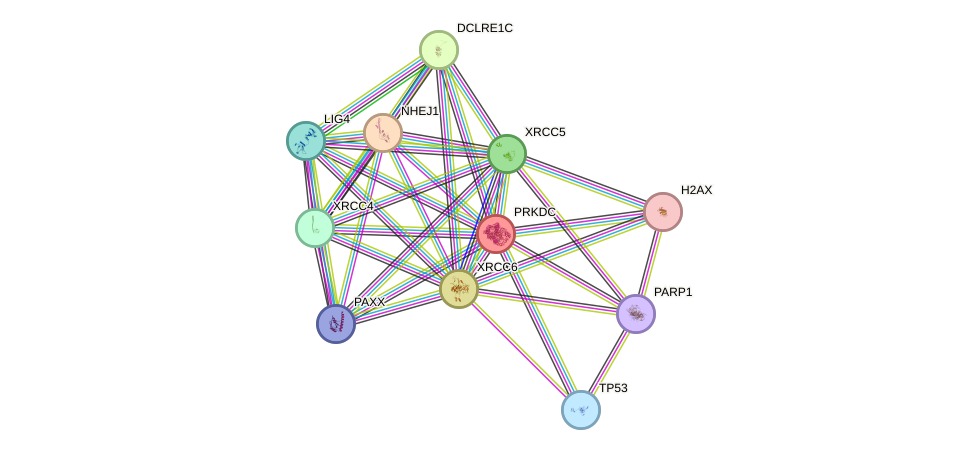
Protein interactions and network
- Protein-protein interacting partners in GenAge
- SHC1, TP53, ATM, WRN, LMNA, HDAC3, AKT1, MYC, EGFR, NBN, PARP1, BRCA1, RPA1, HSP90AA1, NR3C1, ABL1, UBE2I, EP300, GSK3B, TERF1, PRKDC, AR, XRCC5, PCNA, XRCC6, EMD, HSF1, XPA, PRDX1, RELA, MAPK8, SP1, JUN, MAPK9, BMI1, HSPA8, H2AFX, HOXB7, ESR1, SIRT6, MLH1, CHEK2, SIRT7, GSK3A, IKBKB
- STRING interaction network
Retrieve sequences for PRKDC
Homologs in model organisms
- Danio rerio
- wu:fa96e12
- Mus musculus
- Prkdc
- Rattus norvegicus
- Prkdc