GenAge entry for HDAC1 (Homo sapiens)
Gene name (HAGRID: 151)
- HGNC symbol
- HDAC1
- Aliases
- HD1; GON-10; RPD3L1
- Common name
- histone deacetylase 1
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to a pathway or mechanism linked to ageing
- Description
HDAC1 performs the deacetylation of histones and is an important player in may processes such as development, transcriptional regulation, and cell cycle progression [736]. Homologues of HDAC1 have been linked to longevity in yeast [203] and to ageing and caloric restriction in fruit flies [202]. Whether HDAC1 is involved in human ageing, however, is unknown.
Cytogenetic information
- Cytogenetic band
- 1p34
- Location
- 32,292,107 bp to 32,333,623 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0006325; chromatin organization
GO:0006338; chromatin remodeling
GO:0006346; methylation-dependent chromatin silencing
GO:0006351; transcription, DNA-templated
GO:0006476; protein deacetylation
GO:0007596; blood coagulation
GO:0008284; positive regulation of cell proliferation
GO:0009913; epidermal cell differentiation
GO:0010629; negative regulation of gene expression
GO:0010832; negative regulation of myotube differentiation
GO:0010870; positive regulation of receptor biosynthetic process
GO:0016032; viral process
GO:0016575; histone deacetylation
GO:0032922; circadian regulation of gene expression
GO:0042475; odontogenesis of dentin-containing tooth
GO:0042733; embryonic digit morphogenesis
GO:0043044; ATP-dependent chromatin remodeling
GO:0043066; negative regulation of apoptotic process
GO:0043922; negative regulation by host of viral transcription
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0060766; negative regulation of androgen receptor signaling pathway
GO:0060789; hair follicle placode formation
GO:0061029; eyelid development in camera-type eye
GO:0061198; fungiform papilla formation
GO:0070932; histone H3 deacetylation
GO:0070933; histone H4 deacetylation
GO:0090090; negative regulation of canonical Wnt signaling pathway
GO:1901796; regulation of signal transduction by p53 class mediator
GO:1904837; beta-catenin-TCF complex assembly
Cellular component: GO:0000118; histone deacetylase complex
GO:0000785; chromatin
GO:0000790; nuclear chromatin
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005737; cytoplasm
GO:0005829; cytosol
GO:0016580; Sin3 complex
GO:0016581; NuRD complex
GO:0043234; protein complex
Hide GO termsFunction: GO:0000976; transcription regulatory region sequence-specific DNA binding
GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0000980; RNA polymerase II distal enhancer sequence-specific DNA binding
GO:0001047; core promoter binding
GO:0001085; RNA polymerase II transcription factor binding
GO:0001103; RNA polymerase II repressing transcription factor binding
GO:0001106; RNA polymerase II transcription corepressor activity
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0004407; histone deacetylase activity
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0019213; deacetylase activity
GO:0019899; enzyme binding
GO:0031492; nucleosomal DNA binding
GO:0032041; NAD-dependent histone deacetylase activity (H3-K14 specific)
GO:0033558; protein deacetylase activity
GO:0033613; activating transcription factor binding
GO:0042826; histone deacetylase binding
GO:0044212; transcription regulatory region DNA binding
GO:0047485; protein N-terminus binding
GO:0051059; NF-kappaB binding
GO:0070491; repressing transcription factor binding
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, ATM, STAT3, STAT5A, MYC, PARP1, BRCA1, PIN1, CREBBP, HIF1A, TOP2A, TOP2B, NFKB1, UBE2I, ERCC6, EP300, PML, AR, PCNA, RB1, EMD, HSF1, HDAC1, HSPD1, MAPK8, MAPK14, SP1, JUN, MAPK9, CREB1, TBP, APEX1, HBP1, SIN3A, E2F1, HDAC2, MXD1, MDM2, DDIT3, RELA, NR3C1, HELLS, BUB1B, CEBPA, CEBPB, ESR1, NFKBIA, CTNNB1, CDKN2A, PPP1CA, SIRT6, BUB3, ATR, PPARG, NCOR2, TP73, NFE2L2, CDKN1A, PIK3CA
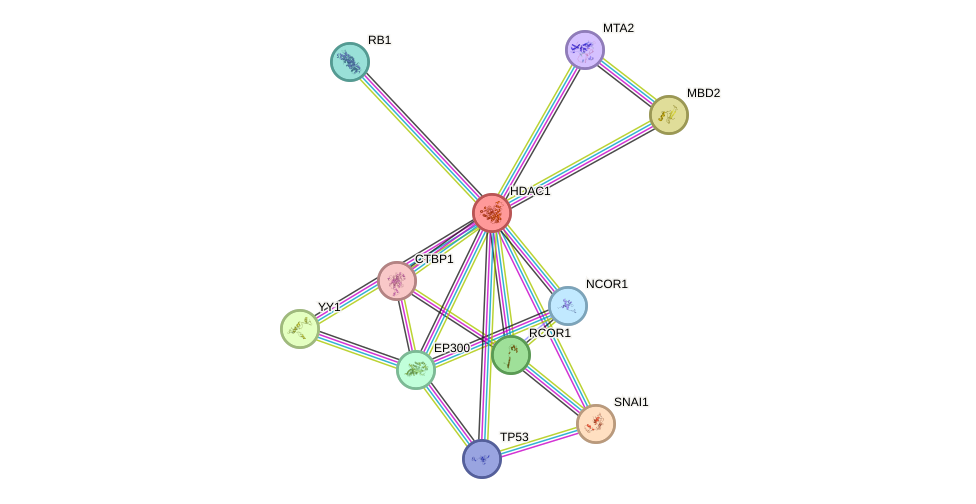
- STRING interaction network
Retrieve sequences for HDAC1
Homologs in model organisms
- Danio rerio
- hdac1
- Drosophila melanogaster
- Rpd3
- Mus musculus
- Hdac1
- Rattus norvegicus
- Hdac1
- Saccharomyces cerevisiae
- RPD3
In other databases
- GenAge model organism genes
- GenDR gene manipulations
- CellAge
- This gene is present as HDAC1