GenAge entry for JUN (Homo sapiens)
Gene name (HAGRID: 172)
- HGNC symbol
- JUN
- Aliases
- c-Jun; AP-1
- Common name
- jun proto-oncogene
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to the regulation or control of genes previously linked to ageing
- Description
JUN is an important transcription factor that is also a member of the AP-1 transcriptional complex. Its signalling pathway has been related to ageing in fruit flies [212] and roundworms [60], though a direct role of JUN has not been demonstrated. Results from mice have shown that AP-1's DNA-binding activity decreases with age [837]. JUN-null mice die at embryonic stages [783]. Further research is needed to assess JUN'1 role in human ageing.
Cytogenetic information
- Cytogenetic band
- 1p32-p31
- Location
- 58,780,791 bp to 58,784,113 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0001525; angiogenesis
GO:0001774; microglial cell activation
GO:0001836; release of cytochrome c from mitochondria
GO:0001889; liver development
GO:0001938; positive regulation of endothelial cell proliferation
GO:0003151; outflow tract morphogenesis
GO:0006366; transcription from RNA polymerase II promoter
GO:0007179; transforming growth factor beta receptor signaling pathway
GO:0007184; SMAD protein import into nucleus
GO:0007265; Ras protein signal transduction
GO:0007568; aging
GO:0007612; learning
GO:0007623; circadian rhythm
GO:0008285; negative regulation of cell proliferation
GO:0009314; response to radiation
GO:0009612; response to mechanical stimulus
GO:0010634; positive regulation of epithelial cell migration
GO:0010941; regulation of cell death
GO:0030224; monocyte differentiation
GO:0031103; axon regeneration
GO:0031953; negative regulation of protein autophosphorylation
GO:0032496; response to lipopolysaccharide
GO:0032870; cellular response to hormone stimulus
GO:0034097; response to cytokine
GO:0035026; leading edge cell differentiation
GO:0035994; response to muscle stretch
GO:0038095; Fc-epsilon receptor signaling pathway
GO:0042127; regulation of cell proliferation
GO:0042493; response to drug
GO:0042542; response to hydrogen peroxide
GO:0043392; negative regulation of DNA binding
GO:0043524; negative regulation of neuron apoptotic process
GO:0043525; positive regulation of neuron apoptotic process
GO:0043547; positive regulation of GTPase activity
GO:0043922; negative regulation by host of viral transcription
GO:0043923; positive regulation by host of viral transcription
GO:0045597; positive regulation of cell differentiation
GO:0045657; positive regulation of monocyte differentiation
GO:0045740; positive regulation of DNA replication
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0048146; positive regulation of fibroblast proliferation
GO:0048661; positive regulation of smooth muscle cell proliferation
GO:0051090; regulation of sequence-specific DNA binding transcription factor activity
GO:0051365; cellular response to potassium ion starvation
GO:0051591; response to cAMP
GO:0051726; regulation of cell cycle
GO:0051899; membrane depolarization
GO:0060395; SMAD protein signal transduction
GO:0061029; eyelid development in camera-type eye
GO:0070374; positive regulation of ERK1 and ERK2 cascade
GO:0071277; cellular response to calcium ion
GO:1902895; positive regulation of pri-miRNA transcription from RNA polymerase II promoter
GO:1990441; negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress
GO:2000144; positive regulation of DNA-templated transcription, initiation
Cellular component: GO:0000228; nuclear chromosome
GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005667; transcription factor complex
GO:0005719; nuclear euchromatin
GO:0005829; cytosol
GO:0017053; transcriptional repressor complex
Hide GO termsFunction: GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0000980; RNA polymerase II distal enhancer sequence-specific DNA binding
GO:0000981; RNA polymerase II transcription factor activity, sequence-specific DNA binding
GO:0000982; transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0001102; RNA polymerase II activating transcription factor binding
GO:0001190; transcriptional activator activity, RNA polymerase II transcription factor binding
GO:0003677; DNA binding
GO:0003682; chromatin binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0003705; transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding
GO:0003713; transcription coactivator activity
GO:0005096; GTPase activator activity
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0019899; enzyme binding
GO:0035497; cAMP response element binding
GO:0042802; identical protein binding
GO:0042803; protein homodimerization activity
GO:0044212; transcription regulatory region DNA binding
GO:0044822; poly(A) RNA binding
GO:0046982; protein heterodimerization activity
GO:0070412; R-SMAD binding
GO:0071837; HMG box domain binding
Protein interactions and network
- Protein-protein interacting partners in GenAge
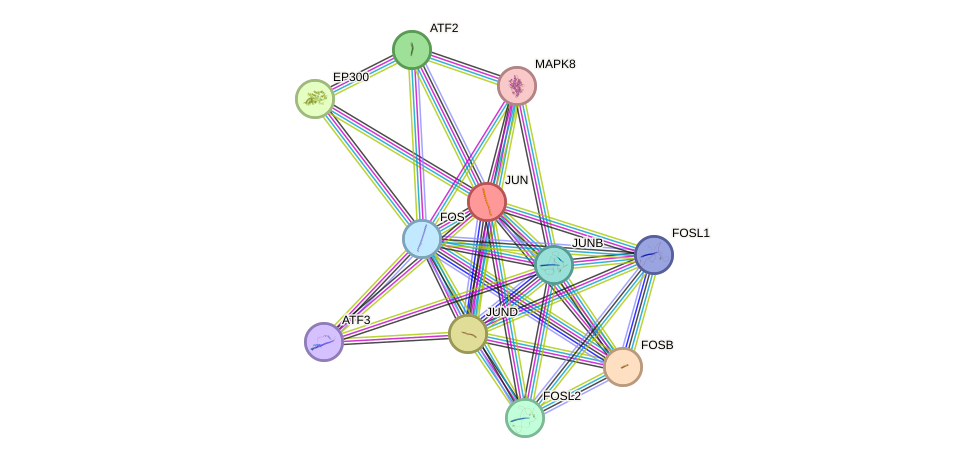
- POU1F1, STAT3, HDAC3, MYC, NCOR1, JUND, FOS, PARP1, BRCA1, PIN1, CREBBP, HIF1A, HSP90AA1, NR3C1, ABL1, TOP2A, UBE2I, CEBPA, CEBPB, EP300, PML, GSK3B, PRKDC, AR, RB1, APP, RELA, SIRT1, HDAC1, MAPK8, MAPK14, SP1, JUN, MAPK9, MAPK3, TAF1, TFAP2A, CREB1, ATF2, TBP, HSPA8, DDIT3, MDM2, SUMO1, ESR1, PPARG, NCOR2, NFE2L2, CDKN1A, IKBKB, NFE2L1
- STRING interaction network
Retrieve sequences for JUN
Homologs in model organisms
In other databases
- CellAge
- This gene is present as JUN