GenAge entry for MYC (Homo sapiens)
Gene name (HAGRID: 39)
- HGNC symbol
- MYC
- Aliases
- c-Myc; bHLHe39; MYCC
- Common name
- v-myc avian myelocytomatosis viral oncogene homolog
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence directly linking the gene product to ageing in a cellular model system
- Description
MYC regulates gene transcription and appears to promote growth. It is also an oncogene [226], involved in cellular senescence [1617]. Overexpression of Myc in flies increases the frequency of somatic mutations, and shortens median and maximum lifespan by up to 47%. Contrastly, Myc haploinsufficiency lowers the frequency of somatic mutations and extends the lifespan of flies by about 14% [3543]. Among its many interacting partners, MYC has been linked to ageing-related genes such as WRN [223] and TERT [225], and hence might play some, so far unknown, role in human ageing.
Cytogenetic information
- Cytogenetic band
- 8q24.21
- Location
- 127,736,069 bp to 127,741,434 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0000165; MAPK cascade
GO:0001658; branching involved in ureteric bud morphogenesis
GO:0002053; positive regulation of mesenchymal cell proliferation
GO:0006112; energy reserve metabolic process
GO:0006338; chromatin remodeling
GO:0006366; transcription from RNA polymerase II promoter
GO:0006879; cellular iron ion homeostasis
GO:0006974; cellular response to DNA damage stimulus
GO:0007050; cell cycle arrest
GO:0007219; Notch signaling pathway
GO:0008284; positive regulation of cell proliferation
GO:0010332; response to gamma radiation
GO:0010468; regulation of gene expression
GO:0010628; positive regulation of gene expression
GO:0015671; oxygen transport
GO:0032204; regulation of telomere maintenance
GO:0032873; negative regulation of stress-activated MAPK cascade
GO:0034644; cellular response to UV
GO:0035690; cellular response to drug
GO:0042493; response to drug
GO:0043066; negative regulation of apoptotic process
GO:0043280; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process
GO:0044346; fibroblast apoptotic process
GO:0045656; negative regulation of monocyte differentiation
GO:0045893; positive regulation of transcription, DNA-templated
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0048146; positive regulation of fibroblast proliferation
GO:0048147; negative regulation of fibroblast proliferation
GO:0050679; positive regulation of epithelial cell proliferation
GO:0051276; chromosome organization
GO:0051782; negative regulation of cell division
GO:0051973; positive regulation of telomerase activity
GO:0060070; canonical Wnt signaling pathway
GO:0070848; response to growth factor
GO:0090096; positive regulation of metanephric cap mesenchymal cell proliferation
GO:1904837; beta-catenin-TCF complex assembly
GO:2000573; positive regulation of DNA biosynthetic process
GO:2001022; positive regulation of response to DNA damage stimulus
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005730; nucleolus
GO:0005829; cytosol
GO:0043234; protein complex
Hide GO termsFunction: GO:0000978; RNA polymerase II core promoter proximal region sequence-specific DNA binding
GO:0001077; transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding
GO:0003677; DNA binding
GO:0003700; transcription factor activity, sequence-specific DNA binding
GO:0005515; protein binding
GO:0008134; transcription factor binding
GO:0032403; protein complex binding
GO:0046983; protein dimerization activity
GO:0070491; repressing transcription factor binding
GO:0070888; E-box binding
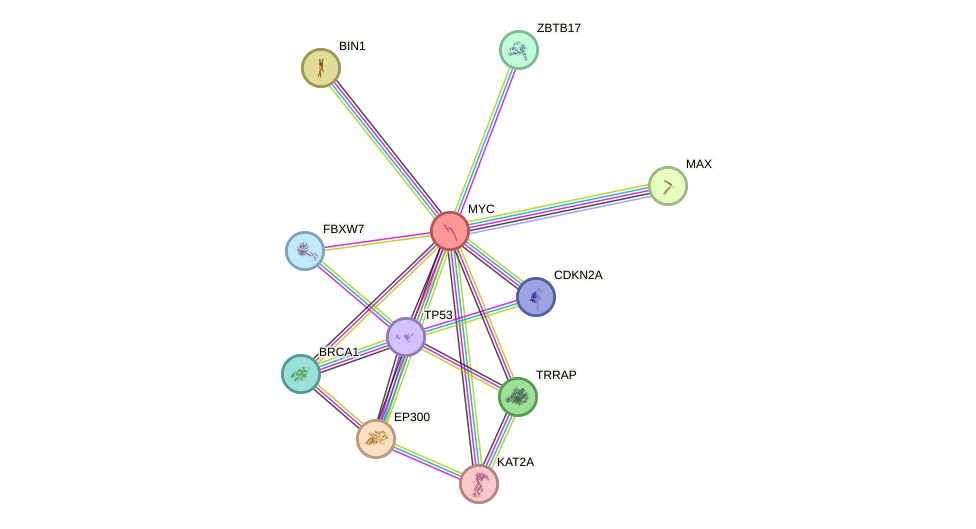
Protein interactions and network
- Protein-protein interacting partners in GenAge
- TP53, PLAU, LMNA, TXN, IRS2, MYC, PRKCD, HDAC3, BRCA1, PIN1, CREBBP, HIF1A, NCOR1, BCL2, HSP90AA1, TOP2A, TOP1, UBE2I, EP300, PML, GSK3B, ERCC3, PRKDC, XRCC5, POLD1, XRCC6, RB1, FOXO3, APP, PRDX1, SIRT1, HDAC1, HSPD1, HSPA1A, MAPK8, SP1, JUN, MED1, MAPK3, TFAP2A, TBP, HBP1, HDAC2, MAX, TAF1, POLA1, RELA, HELLS, CEBPA, CEBPB, ESR1, CDKN2A, PPP1CA, MLH1, CSNK1E, STUB1, TP73, NFE2L2, GSK3A, SQSTM1
- STRING interaction network
Retrieve sequences for MYC
Homologs in model organisms
In other databases
- GenAge model organism genes
- A homolog of this gene for Mus musculus is present as Myc
- CellAge
- This gene is present as MYC