GenAge entry for APP (Homo sapiens)
Gene name (HAGRID: 137)
- HGNC symbol
- APP
- Aliases
- AD1
- Common name
- amyloid beta (A4) precursor protein
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on indirect or inconclusive evidence linking the gene product to ageing in humans or in one or more model systems
- Description
APP has different functions. It is a cell surface receptor, interacts with many other proteins, and may regulate neurite growth. Clearly, APP is an important player in Alzheimer's disease [695], though it is not known whether it is related to other aspects of ageing.
Cytogenetic information
- Cytogenetic band
- 21q21.3
- Location
- 25,880,550 bp to 26,170,820 bp
- Orientation
- Minus strand
Protein information
- Gene Ontology
-
Process: GO:0001967; suckling behavior
GO:0002576; platelet degranulation
GO:0006378; mRNA polyadenylation
GO:0006417; regulation of translation
GO:0006468; protein phosphorylation
GO:0006878; cellular copper ion homeostasis
GO:0006897; endocytosis
GO:0006979; response to oxidative stress
GO:0007155; cell adhesion
GO:0007176; regulation of epidermal growth factor-activated receptor activity
GO:0007219; Notch signaling pathway
GO:0007409; axonogenesis
GO:0007617; mating behavior
GO:0007626; locomotory behavior
GO:0008088; axo-dendritic transport
GO:0008203; cholesterol metabolic process
GO:0008344; adult locomotory behavior
GO:0008542; visual learning
GO:0009987; cellular process
GO:0010288; response to lead ion
GO:0010951; negative regulation of endopeptidase activity
GO:0010952; positive regulation of peptidase activity
GO:0010971; positive regulation of G2/M transition of mitotic cell cycle
GO:0016199; axon midline choice point recognition
GO:0016322; neuron remodeling
GO:0016358; dendrite development
GO:0030198; extracellular matrix organization
GO:0030900; forebrain development
GO:0031175; neuron projection development
GO:0035235; ionotropic glutamate receptor signaling pathway
GO:0040014; regulation of multicellular organism growth
GO:0043393; regulation of protein binding
GO:0044267; cellular protein metabolic process
GO:0045087; innate immune response
GO:0045665; negative regulation of neuron differentiation
GO:0045931; positive regulation of mitotic cell cycle
GO:0045944; positive regulation of transcription from RNA polymerase II promoter
GO:0048669; collateral sprouting in absence of injury
GO:0050803; regulation of synapse structure or activity
GO:0050885; neuromuscular process controlling balance
GO:0051124; synaptic growth at neuromuscular junction
GO:0051402; neuron apoptotic process
GO:0051563; smooth endoplasmic reticulum calcium ion homeostasis
GO:0071320; cellular response to cAMP
GO:0071874; cellular response to norepinephrine stimulus
GO:1990000; amyloid fibril formation
GO:1990090; cellular response to nerve growth factor stimulus
Cellular component: GO:0005576; extracellular region
GO:0005615; extracellular space
GO:0005641; nuclear envelope lumen
GO:0005737; cytoplasm
GO:0005768; endosome
GO:0005790; smooth endoplasmic reticulum
GO:0005791; rough endoplasmic reticulum
GO:0005794; Golgi apparatus
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0005887; integral component of plasma membrane
GO:0005905; clathrin-coated pit
GO:0005911; cell-cell junction
GO:0009986; cell surface
GO:0016021; integral component of membrane
GO:0030134; ER to Golgi transport vesicle
GO:0030424; axon
GO:0031093; platelet alpha granule lumen
GO:0031594; neuromuscular junction
GO:0031904; endosome lumen
GO:0032588; trans-Golgi network membrane
GO:0035253; ciliary rootlet
GO:0043195; terminal bouton
GO:0043197; dendritic spine
GO:0043198; dendritic shaft
GO:0043231; intracellular membrane-bounded organelle
GO:0043235; receptor complex
GO:0044304; main axon
GO:0045121; membrane raft
GO:0045177; apical part of cell
GO:0045202; synapse
GO:0048471; perinuclear region of cytoplasm
GO:0051233; spindle midzone
GO:0070062; extracellular exosome
GO:0097449; astrocyte projection
GO:1990761; growth cone lamellipodium
GO:1990812; growth cone filopodium
Hide GO termsFunction: GO:0003677; DNA binding
GO:0004867; serine-type endopeptidase inhibitor activity
GO:0005102; receptor binding
GO:0005515; protein binding
GO:0008201; heparin binding
GO:0016504; peptidase activator activity
GO:0019899; enzyme binding
GO:0042802; identical protein binding
GO:0046914; transition metal ion binding
GO:0051425; PTB domain binding
GO:0070851; growth factor receptor binding
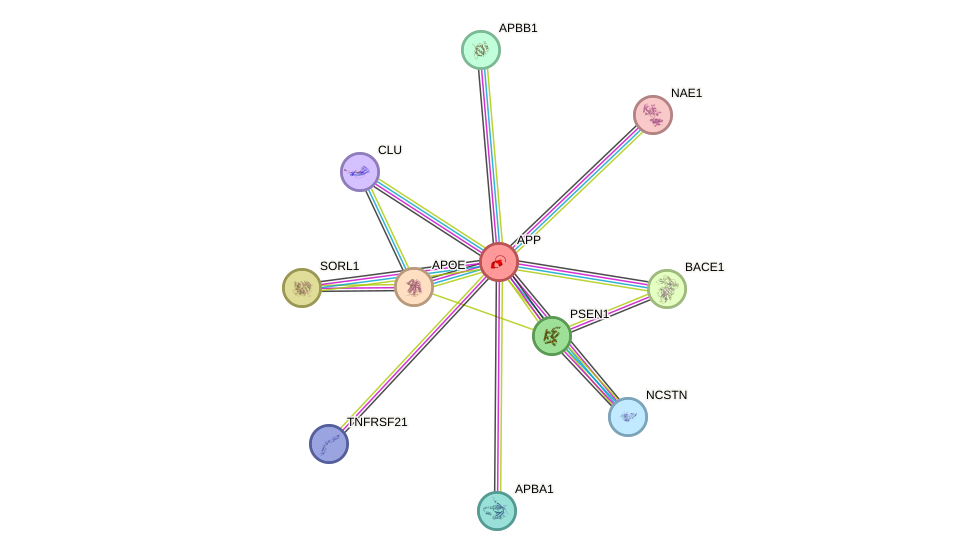
Protein interactions and network
- Protein-protein interacting partners in GenAge
- SHC1, IGF1R, STAT5B, INS, IRS1, AKT1, MYC, EGFR, FOS, PRKCD, HSP90AA1, TOP1, UBE2I, TGFB1, STK11, GSK3B, CAT, FEN1, HSPA8, SOD2, LRP2, UCHL1, APP, A2M, ATP5O, ERCC1, HSPD1, MAPK8, YWHAZ, PTK2, IL7, JUN, HMGB2, MAP3K5, BDNF, ATF2, APEX1, PTGS2, HSPA9, MXD1, MDM2, UBB, MXI1, HOXB7, HOXC4, HELLS, CLU, CTNNB1, PSEN1, MAPT, PDPK1, CSNK1E, STUB1, CHEK2, PCK1, PPARG, CISD2, SIRT7, CDKN1A, CDKN2B, CDK7, CTF1, TRAP1
- STRING interaction network
Retrieve sequences for APP
Homologs in model organisms
In other databases
- CellAge gene expression
- This gene is present as APP