GenAge entry for MDM2 (Homo sapiens)
Gene name (HAGRID: 210)
- HGNC symbol
- MDM2
- Aliases
- HDM2; MGC5370
- Common name
- MDM2 proto-oncogene, E3 ubiquitin protein ligase
Potential relevance to the human ageing process
- Main reason for selection
- Entry selected based on evidence linking the gene product to the regulation or control of genes previously linked to ageing
- Description
MDM2 is an oncogene that inhibits TP53 [890]. The MDM2 gene encodes multiple transcripts, many of which are tissue-specific. Disruption of MDM2 in mice resulted in increased TP53-dependent apoptosis and defects in multiple haematopoietic lineages [889]. Mutations in the human MDM2 gene have been associated with cancer [1738]. A role for MDM2 in human ageing is unknown.
Cytogenetic information
- Cytogenetic band
- 12q14.3-q1
- Location
- 68,808,172 bp to 68,845,544 bp
- Orientation
- Plus strand
Protein information
- Gene Ontology
-
Process: GO:0000122; negative regulation of transcription from RNA polymerase II promoter
GO:0001568; blood vessel development
GO:0001974; blood vessel remodeling
GO:0002027; regulation of heart rate
GO:0003181; atrioventricular valve morphogenesis
GO:0003203; endocardial cushion morphogenesis
GO:0003281; ventricular septum development
GO:0003283; atrial septum development
GO:0006461; protein complex assembly
GO:0006977; DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest
GO:0007089; traversing start control point of mitotic cell cycle
GO:0008284; positive regulation of cell proliferation
GO:0009636; response to toxic substance
GO:0010039; response to iron ion
GO:0010628; positive regulation of gene expression
GO:0010955; negative regulation of protein processing
GO:0010977; negative regulation of neuron projection development
GO:0016032; viral process
GO:0016567; protein ubiquitination
GO:0016925; protein sumoylation
GO:0018205; peptidyl-lysine modification
GO:0031648; protein destabilization
GO:0032026; response to magnesium ion
GO:0032436; positive regulation of proteasomal ubiquitin-dependent protein catabolic process
GO:0034504; protein localization to nucleus
GO:0036369; transcription factor catabolic process
GO:0042176; regulation of protein catabolic process
GO:0042220; response to cocaine
GO:0042493; response to drug
GO:0042787; protein ubiquitination involved in ubiquitin-dependent protein catabolic process
GO:0043154; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process
GO:0043278; response to morphine
GO:0043518; negative regulation of DNA damage response, signal transduction by p53 class mediator
GO:0045184; establishment of protein localization
GO:0045472; response to ether
GO:0045892; negative regulation of transcription, DNA-templated
GO:0045931; positive regulation of mitotic cell cycle
GO:0046677; response to antibiotic
GO:0046827; positive regulation of protein export from nucleus
GO:0048545; response to steroid hormone
GO:0051865; protein autoubiquitination
GO:0060411; cardiac septum morphogenesis
GO:0070301; cellular response to hydrogen peroxide
GO:0071157; negative regulation of cell cycle arrest
GO:0071236; cellular response to antibiotic
GO:0071301; cellular response to vitamin B1
GO:0071312; cellular response to alkaloid
GO:0071363; cellular response to growth factor stimulus
GO:0071375; cellular response to peptide hormone stimulus
GO:0071391; cellular response to estrogen stimulus
GO:0071456; cellular response to hypoxia
GO:0071494; cellular response to UV-C
GO:1901796; regulation of signal transduction by p53 class mediator
GO:1901797; negative regulation of signal transduction by p53 class mediator
GO:1904404; response to formaldehyde
GO:1904707; positive regulation of vascular smooth muscle cell proliferation
GO:1904754; positive regulation of vascular associated smooth muscle cell migration
GO:1990785; response to water-immersion restraint stress
Cellular component: GO:0005634; nucleus
GO:0005654; nucleoplasm
GO:0005730; nucleolus
GO:0005737; cytoplasm
GO:0005829; cytosol
GO:0005886; plasma membrane
GO:0016604; nuclear body
GO:0030666; endocytic vesicle membrane
GO:0043234; protein complex
GO:0045202; synapse
Hide GO termsFunction: GO:0002039; p53 binding
GO:0004842; ubiquitin-protein transferase activity
GO:0005515; protein binding
GO:0008270; zinc ion binding
GO:0016874; ligase activity
GO:0019789; SUMO transferase activity
GO:0019899; enzyme binding
GO:0031625; ubiquitin protein ligase binding
GO:0042802; identical protein binding
GO:0042975; peroxisome proliferator activated receptor binding
GO:0061630; ubiquitin protein ligase activity
GO:0097110; scaffold protein binding
Protein interactions and network
- Protein-protein interacting partners in GenAge
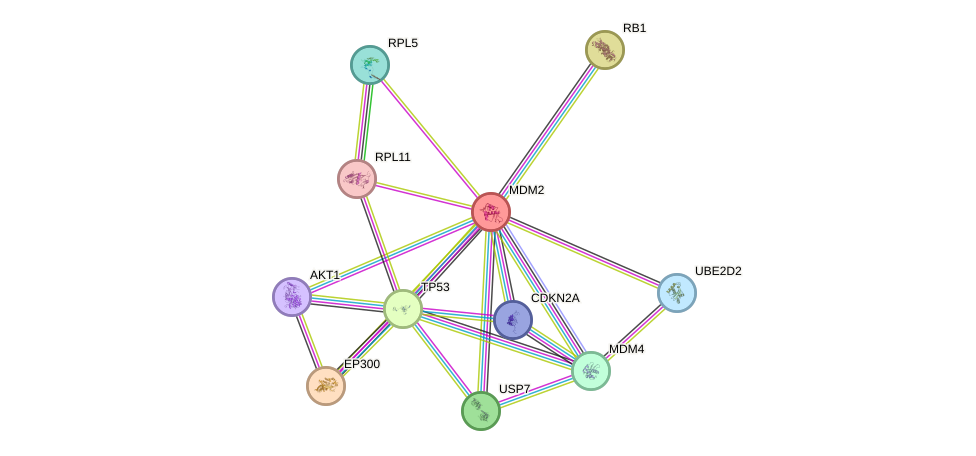
- TP53, TERT, ATM, ERCC8, IGF1R, E2F1, AKT1, NBN, JUND, CREBBP, HIF1A, S100B, VCP, HSP90AA1, NR3C1, EGR1, ABL1, TOP1, UBE2I, ERCC6, EP300, PML, GSK3B, EEF2, AR, PCNA, XRCC6, RB1, FOXO3, FOXO1, UCHL1, APP, RELA, HDAC1, PTK2, JUN, MED1, TAF1, SDHC, FOXO4, EEF1A1, TBP, APEX1, BUB1B, HSPA8, MDM2, SUMO1, ESR1, CLU, CDKN2A, PPP1CA, TP63, SIRT6, PPM1D, CHEK2, PPARG, TP73, CDKN1A
- STRING interaction network
Retrieve sequences for MDM2
Homologs in model organisms
In other databases
- LongevityMap
- This gene is present as MDM2
- CellAge
- This gene is present as MDM2
- CellAge gene expression
- This gene is present as MDM2